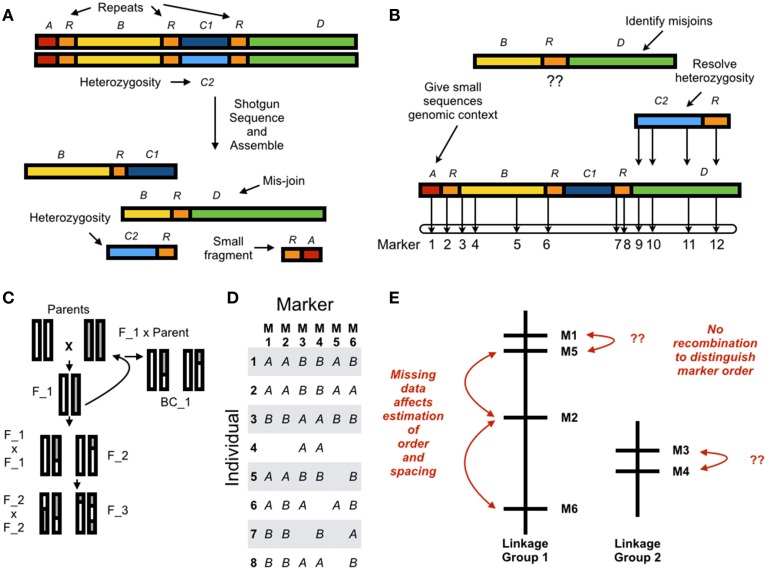
Figure 1.
(A) In whole genome assembly errors result from residual alleles which appear as discrete sequences in the reference, and mis-joins. Small fragments have no genomic context and contribute little information. (B) Using a genetic linkage map to anchor a de novo assembly resolves error in the reference sequence by giving small sequences genomic context, resolving allelism, and identifying mis-joins. Chromosome-scale assemblies can be constructed by ordering and orienting sequences with the linkage map. (C) A genetic linkage map can be estimated from a parental cross resulting in an F2, F3, or Backcross (here, BC1) population. Estimating a genetic linkage map requires (D) genotyping individuals at discrete markers (here, six markers across eight individuals with missing data); and (E) grouping markers into linkage groups; and ordering and spacing markers within linkage groups. Estimating order and spacing is difficult due to missing data and little recombination between adjacent markers.