Abstract
Mixed infections and heteroresistance of Mycobacterium tuberculosis contribute to the difficulty of diagnosis, treatment, and control of tuberculosis. However, there is still no proper solution for these issues. This study aimed to investigate the potential relationship between mixed infections and heteroresistance and to determine the high-risk groups related to these factors. A total of 499 resistant and susceptible isolates were subjected to spoligotyping and 24-locus variable-number tandem repeat methods to analyze their genotypic lineages and the occurrence of mixed infections. Two hundred ninety-two randomly selected isolates were sequenced on their rpoB gene to examine mutations and heteroresistance. The results showed that 12 patients had mixed infections, and the corresponding isolates belonged to Manu2 (n = 8), Beijing (n = 2), T (n = 1), and unknown (n = 1) lineages. Manu2 was found to be significantly associated with mixed infections (odds ratio, 47.72; confidence interval, 9.68 to 235.23; P < 0.01). Four isolates (1.37%) were confirmed to be heteroresistant, which was caused by mixed infections in three (75%) isolates; these belonged to Manu2. Additionally, 3.8% of the rifampin-resistant isolates showing no mutation in the rpoB gene were significantly associated with mixed infections (χ2, 56.78; P < 0.01). This study revealed for the first time that Manu2 was the predominant group in the cases of mixed infections, and this might be the main reason for heteroresistance and a possible mechanism for isolates without any mutation in the rpoB gene to become rifampin resistant. Further studies should focus on this lineage to clarify its relevance to mixed infections.
INTRODUCTION
Mixed Mycobacterium tuberculosis infections were recognized as early as 1975 (1), and they continue to cause an increase in the false-negative rate of drug resistance testing obtained by both drug susceptibility tests (DST) and GeneXpert (2–4). Mixed infections can assist M. tuberculosis strains in acquiring additional mutations, facilitate the spread of drug-resistant strains, and boost the rate of treatment failure (3, 5). With the popularization of genetic analysis tools, the situation in which a single patient gets infected with more than one M. tuberculosis isolate was found to be more common than we previously expected (6). Currently, mycobacterial interspersed repetitive unit-variable-number tandem repeat (MIRU-VNTR)-based methods have been the most widely used to detect mixed infections (5). Mixed infections are defined by the presence of strains with different MIRU-VNTR patterns at two or more loci in the same sputum, lymph, or other sample while having clonal heterogeneity with a different MIRU-VNTR pattern at a single locus (7–9).
In some cases, quickly acquired drug resistance may be caused by misdiagnosis of mixed infections or heteroresistance (3, 10, 11). However, there are only very few publications on heteroresistance (11), which is defined as the coexistence of susceptible and resistant bacteria in the same sample (12). Heteroresistance, as another confounding factor in diagnosis and treatment, is more difficult to detect by the phenotypic DST method from the preliminary stage to full resistance (11, 12). In comparison, DNA sequencing, a robust method for analyzing sequence variants, with results showing dual peaks in the rpoB gene region, can be regarded as the gold standard to determine rifampin heteroresistance (11). There are two mechanisms for heteroresistance, one being the mixed infection, and the other being the splitting of a single strain into susceptible and resistant clones due to evolution.
The diagnosis of mixed infection is expensive and labor-intensive, since it requires the lineages of single colonies to be identified individually from the original culture (6). In the cases of mixed infections, the usefulness of spoligotyping might be compromised, as a spoligotype pattern may reflect the cumulative spacers of all strains present or that of the dominant strain (7). However, if we could confirm certain spoligotypes whose existence was most probably related to mixed infection, it would be easier to investigate and control this situation (6). A computer program has been designed to investigate some “troublesome” spoligotype lineages by generating relative occurrences of one spoligotype being the combination of two known spoligotypes, but the results still need to be confirmed by MIRU-VNTR or other methods (6). To date, only one study using an in-house PCR-based method to detect some defined lineages was able to determine whether certain strain lineages were prone to cause mixed infections (10). However, this method did not determine the extent of mixed infections with different strains of the same lineage or other undefined strain lineages whose primers were not included, such as Manu.
Mixed infections and heteroresistance may be particularly common in regions with a high rate of tuberculosis (TB), especially drug-resistant TB (5, 6, 11). Sichuan is the province with the second-largest number of TB cases in China, where the prevalence of drug-resistant TB is much higher than the average level observed in eastern China (13). Therefore, the objectives of this study were to map the prevalence of mixed infections and heteroresistance of M. tuberculosis in Sichuan and their potential correlation, and to investigate the group with a high risk of mixed infections and heteroresistance.
MATERIALS AND METHODS
Study design and isolates.
A retrospective cross-section study was conducted between January 2008 and March 2011. A total of 5,090 M. tuberculosis clinical isolates, including 3,356 (65.9%) pansusceptible isolates and 1,734 (34.1%) drug-resistant isolates, were collected from 5,090 pulmonary tuberculosis patients coming mainly from Sichuan Province. These patients were diagnosed at the Chengdu Public Health Clinical Center, the only professional antituberculosis hospital in Sichuan. Among the 1,734 resistant isolates, 415 were randomly selected, 82 isolates from the 3,356 pansusceptible isolates were selected, and 2 isolates were without any clinical information were selected; in total, 499 isolates were selected, with one isolate per patient.
Clinical data, including treatment time, gender, age, and nationality, were obtained from the medical records of the subjects without collecting private information about the patients or disclosing information about the patients to any commercial agency. This study was approved by the Medical Ethics Committee of Chengdu Public Health Clinical Center. All patients included in this study provided written informed consent for the use of clinical samples. The results of this study did not influence patient treatment in any way.
Drug susceptibility.
Samples were collected and disposed of in accordance with WHO guidelines. Briefly, strains were cultured on Lowenstein-Jensen (LJ) slants at 37°C, and M. tuberculosis complex isolates were identified using standard biochemical methods, such as susceptibility to p-nitrobenzoic acid (PNB) and 2-thiopnene carboxylic acid hydrazide (TCH), pyrazinamidase activity (PZA), nitrate reduction, and niacin production. DST was done using a proportion method with 10 mg/ml streptomycin (STR), 0.2 mg/ml isoniazid (INH), 40 mg/ml rifampin (RIF), and 2 mg/ml ethambutol (EMB), as described elsewhere (2, 14, 15). The DST was repeated for all samples showing discordant results between phenotypic DST and mutation analysis.
DNA extract and genotyping.
Genomic DNA from clinical isolates was extracted using the cetyltrimethylammonium bromide (CTAB) method (16). Spoligotyping was performed as described by Kamerbeek et al. (17). For MIRU-VNTRs, all 24 loci were amplified with the corresponding primers, as described by Supply et al. (18).
To avoid laboratory cross-contamination, standard precautions included physical separation of pre- and post-PCR areas, a separate environment control, and unidirectional workflow. Other precautionary measures included the use of dedicated consumables and equipment (including aerosol-resistant pipettes, use of the proper pipetting technique, and fresh gloves used in a PCR area), aseptic cleaning carried out periodically before and after PCR work, and periodic wipe tests. Lastly, only a limited number of samples were processed at one time, and DNA from M. tuberculosis strain H37Rv and deionized water was added as positive and negative controls, respectively, for each PCR. The PCR was repeated three times if the sample showed more than one band at any MIRU-VNTR locus in order to confirm the result.
PCR of rpoB and sequencing.
PCR procedures and two oligonucleotide primers for rpoB were used as described by Tang et al. (19). The amplicons were purified with QIAquick PCR purification kits (Qiagen, United Kingdom), according to the manufacturer's instructions. Direct sequencing of the PCR products was carried out with an ABI Prism 377 automated DNA sequencer (Applied Biosystems) and BigDye Terminator cycle sequencing kit (ABI Prism), according to the instructions provided by the manufacturer. The sequencing process was repeated three times to confirm the results from patients with mixed infections or heteroresistance.
Statistical analysis.
The analysis of PCR fragment size and the assignment of the various VNTR alleles were achieved using the Quantity One (version 4.6.2) software (Bio-Rad Laboratories). The Hunter-Gaston discriminatory index (HGDI) was calculated as described previously (20). The spoligotypes and MIRU-VNTR patterns were compared using the SITVIT2 proprietary database of the Institut Pasteur de la Guadeloupe, which is an updated version of the previously released SITVIT WEB database (21) (see http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/).
Univariate logistic regression analysis was performed to identify potential influencing factors from patient and bacterial variables associated with mixed infections. Odds ratios (OR) and 95% confidence intervals (CI) were examined. A P value of <0.05 was considered statistically significant. Pearson's chi-square exact test (two-tailed) was used to compare the differences. The data were analyzed using the Stata statistical software (version 12; Stata Corporation, College Town, TX, USA).
The sequences were analyzed for the presence of single-nucleotide polymorphisms using the Phred/Phrap/PolyPhred/Consed software package (http://droog.gs.washington.edu/PolyPhred.html). The analytical results of rpoB sequencing were compared with M. tuberculosis H37Rv (GenBank accession no. NC_000962).
RESULTS
Diversity of M. tuberculosis by spoligotyping.
Based on the spoligotyping results, 22 orphan patterns and 61 shared types (n = 477 isolates) were distributed in seven lineages from a total of 499 isolates of M. tuberculosis in Sichuan, China (Table 1). The Beijing family was the predominant group, representing 69.74% (n = 348) of all isolates, followed by the T family (n = 83 [16.63%]) and Manu2 (n = 37 [7.41%]). Eighteen (3.61%) isolates showed unknown patterns that were not assigned to any known major lineages in the SITVIT2 database, and 13 (2.61%) belonged to other minor lineages. Among the 22 orphan patterns, 20 (90.9%) belonged to unknown (n = 5 [33.33%]), T (n = 7 [8.43%]), and Manu2 (n = 8 [21.62%]) lineages.
TABLE 1.
Distribution of mixed infections in 61 SITs (n = 477 isolates) and 22 orphan isolates with corresponding genotypes defined by spoligotypinga
| SIT | Spoligotype description | Octal no. | Total no. (%) of isolates in this study | % in study vs database | No. (% in the SIT) of mixed infections in this study | Clade |
|---|---|---|---|---|---|---|
| 1 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■■■■■ | 000000000003771 | 324 (64.93) | 3.11 | 2 (0.62) | Beijing |
| 26 | ■■■□□□□■■■■■■■■■■■■■■■□□□□□□□□□□□□■■■■■■■■■ | 703777740003771 | 1 (0.2) | 0.06 | 0 (0) | CAS1-Delhi |
| 37 | ■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777737777760771 | 1 (0.2) | 0.19 | 0 (0) | T3 |
| 42 | ■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■■□□□□■■■■■■■ | 777777607760771 | 1 (0.2) | 0.03 | 0 (0) | LAM9 |
| 44 | ■■■■■■■■■■■■■■■■■■■■■■□■■■■■■■■■□□□□■■■■■■■ | 777777757760771 | 1 (0.2) | 0.46 | 0 (0) | T5 |
| 50 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□■□□□□■■■■■■■ | 777777777720771 | 3 (0.6) | 0.08 | 0 (0) | H3 |
| 52 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 777777777760731 | 10 (2) | 1.05 | 0 (0) | T2 |
| 53 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777777777760771 | 13 (2.61) | 0.2 | 0 (0) | T1 |
| 54 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | 777777777763771 | 12 (2.4) | 4.53 | 4 (33.33) | Manu2 |
| 118 | ■■■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777767777760771 | 1 (0.2) | 0.61 | 0 (0) | T1 |
| 154 | ■■■■□■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 757777777760771 | 1 (0.2) | 0.88 | 0 (0) | T1 |
| 159 | ■■■■■■■■■■■■■□□□□□□□□□□■■■■■■■■■□□□□■■■■■■■ | 777740017760771 | 1 (0.2) | 1.32 | 0 (0) | Unknown |
| 189 | ■■■■■■■■■■■■■□□□□■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777741777760771 | 1 (0.2) | 7.69 | 0 (0) | T1 |
| 190 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■□■■■ | 000000000003731 | 4 (0.8) | 2.17 | 0 (0) | Beijing |
| 191 | □□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 177777777760771 | 1 (0.2) | 4.76 | 0 (0) | T1 |
| 196 | ■■□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 677777777760771 | 1 (0.2) | 1.2 | 0 (0) | T1 |
| 250 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■■ | 000000000000371 | 1 (0.2) | 2.63 | 0 (0) | Beijing |
| 265 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■□■■■■■■ | 000000000003371 | 1 (0.2) | 1.23 | 0 (0) | Beijing |
| 269 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■■■ | 000000000000771 | 1 (0.2) | 1.08 | 0 (0) | Beijing |
| 278 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■□■ | 777777777760761 | 1 (0.2) | 8.33 | 0 (0) | T1 |
| 291 | ■■■■■■■■■■■■■■■■■■■■□■■■■■■■■■■■□□□□■■■■■■■ | 777777677760771 | 1 (0.2) | 1.11 | 0 (0) | T1 |
| 294 | ■□■■■■■■■■■■■■■■■■■■■■■■■■■■■■□■□□□□■■■■■■■ | 577777777720771 | 3 (0.6) | 9.09 | 0 (0) | H3 |
| 334 | ■□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 577777777760771 | 18 (3.61) | 16.22 | 0 (0) | T1 |
| 393 | ■■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777757777760771 | 4 (0.8) | 10 | 1 (25) | T1 |
| 462 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■□■■■□□□□■■■■■■■ | 777777777560771 | 1 (0.2) | 1.72 | 0 (0) | T1 |
| 511 | ■■■■■■■■■■■■■■■■■■■■■□□□□□□□□□□■□□□□■■■■■■■ | 777777700020771 | 2 (0.4) | 4.26 | 0 (0) | H3 |
| 583 | ■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | 777737777763771 | 1 (0.2) | 33.33 | 0 (0) | Manu2 |
| 616 | ■□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■■■■■ | 400000000003771 | 1 (0.2) | 9.09 | 0 (0) | Unknown |
| 621 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■□■■■■■■■ | 000000000002771 | 3 (0.6) | 8.33 | 0 (0) | Beijing |
| 632 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■□■■■■■ | 000000000003571 | 6 (1.2) | 20.69 | 0 (0) | Beijing |
| 780 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□□□□■■■■■■■ | 777777777600771 | 1 (0.2) | 7.14 | 0 (0) | Unknown |
| 796 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■■■■ | 000000000001771 | 1 (0.2) | 9.09 | 0 (0) | Beijing |
| 803 | ■■■■■■■■■■■■■□□□□□□□□□□□■■■■■■■■□□□□■■■■■■■ | 777740007760771 | 1 (0.2) | 2.94 | 0 (0) | LAM9 |
| 804 | ■□□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 477777777760771 | 1 (0.2) | 3.85 | 0 (0) | T1 |
| 834 | ■■■■■■■■■■■■■■□■■■■■■■■■■■■■■■■□□□□□■■■■■■■ | 777767777740771 | 1 (0.2) | 12.5 | 0 (0) | T1 |
| 848 | ■■■□■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 737777777760731 | 1 (0.2) | 3.23 | 0 (0) | T2 |
| 1096 | ■□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | 577777777763771 | 15 (3.01) | 66.67 | 3 (20) | Manu2 |
| 1122 | ■■■■■□■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 767777777760771 | 1 (0.2) | 1.67 | 0 (0) | T1 |
| 1168 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■□□■■■ | 000000000003631 | 1 (0.2) | 14.29 | 0 (0) | Beijing |
| 1302 | ■□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 577777777760731 | 4 (0.8) | 16.67 | 0 (0) | T2 |
| 1451 | ■■■■■■■■■■■■■■□□□□□□□□□□□□□□□□□□□□□□□□□□□□□ | 777760000000000 | 1 (0.2) | 7.14 | 0 (0) | Unknown |
| 1475 | ■■■■■■■■■□■■■□■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777357777760771 | 1 (0.2) | 11.11 | 0 (0) | T1 |
| 1578 | ■■■■■□■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 767777777760731 | 1 (0.2) | 14.29 | 0 (0) | T2 |
| 1674 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■■■□■ | 000000000003761 | 1 (0.2) | 14.29 | 0 (0) | Beijing |
| 1690 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□■□■■■■■■■ | 777777777762771 | 1 (0.2) | 25 | 0 (0) | Manu2 |
| 1890 | ■■■■■■■■■■■■■■■■■■■■■□□□■■■■■■■■□□□□■■■□■■■ | 777777707760731 | 1 (0.2) | 9.09 | 0 (0) | T2 |
| 2276 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□■■■■■■■■■ | 777777777743771 | 2 (0.4) | 50 | 1 (50) | Unknown |
| 2979 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■ | 000000000000171 | 2 (0.4) | 22.22 | 0 (0) | Beijing |
| 3199 | ■■■■■■□□■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 771777777760731 | 1 (0.2) | 33.33 | 0 (0) | T2 |
| 3233 | ■■■□□□□□□□■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 700377777760731 | 3 (0.6) | 100 | 0 (0) | T2 |
| 3234 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□□□□□■■■■■■ | 777777777600371 | 2 (0.4) | 50 | 0 (0) | Unknown |
| 3235 | ■■□□□□■■■■■■■■■■■■■■■■■□□□□□□□■■□□□□■■■□■■■ | 607777760060731 | 2 (0.4) | 100 | 0 (0) | T2 |
| 3236 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■□□□□□■■■ | 000000000002031 | 1 (0.2) | 50 | 0 (0) | Beijing |
| 3237 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□□□□□□□□□□■■ | 777777777400011 | 1 (0.2) | 50 | 0 (0) | Unknown |
| 3238 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□□□■■□□□□ | 777777777740300 | 2 (0.4) | 100 | 0 (0) | T1 |
| 3239 | ■■■■■■■■■■■■■■□□□□□□□□□□□□□□□□□□□□□□■■■□■■■ | 777760000000731 | 1 (0.2) | 50 | 0 (0) | Unknown |
| 3240 | ■■■■■■■■□□□■■■□■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 776167777760731 | 1 (0.2) | 50 | 0 (0) | T2 |
| 3241 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■□□□□□ | 000000000003600 | 2 (0.4) | 50 | 0 (0) | Beijing |
| 3309* | ■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■■■□□□□■□■■■■■ | 777737777760571 | 1 (0.2) | 20 | 0 (0) | T2-Uganda |
| 4011* | □□□□□□□□□□□□□□□□□□□■□□□□□□□□□□□□□□■■■■■■■■■ | 000000200003771 | 2 (0.4) | 100 | 0 (0) | Unknown |
| 4012* | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■■■ | 777777777703771 | 1 (0.2) | 50 | 0 (0) | Unknown |
| Orphan | ■■■■■■■■■■■■■■■■■■■■■□□□■□□□■■□□□□■■■■■■■■■ | 777777704303771 | 1 (0.2) | 100 | 0 (0) | Unknown |
| Orphan | ■■□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■ | 600000000000011 | 1 (0.2) | 100 | 0 (0) | Mungi |
| Orphan | ■■■■■■■■■■■■■□□□□□□□□□□□■■■■■■■■□□□□■□■■■■■ | 777740007760571 | 1 (0.2) | 100 | 0 (0) | LAM9 |
| Orphan | ■■□■■□■■■■■■■■■■■■■■■■■■■■□□■■■■□□□□■■■■■■■ | 667777776360771 | 1 (0.2) | 100 | 0 (0) | T1 |
| Orphan | ■■■■■□■■■■■■■□□□□□□□□□□□□■■■■■■■□□□□■■■■■■■ | 767740003760771 | 1 (0.2) | 100 | 0 (0) | T1 |
| Orphan | ■■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■■□□□□■□■□■■■ | 777757777760531 | 1 (0.2) | 100 | 0 (0) | T2 |
| Orphan | ■□□■■■■■■■□□■■■■■■■■■■■■■■■■■■■■□□□□■■■□■■■ | 477477777760731 | 1 (0.2) | 100 | 0 (0) | T2 |
| Orphan | ■□□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | 477777777763771 | 1 (0.2) | 100 | 1 (100) | Manu2 |
| Orphan | ■□■■■■■■□□□■■■■□□■■■■■■■■■■■■■■■□□□□■■■□■■■ | 576171777760731 | 1 (0.2) | 100 | 0 (0) | T2 |
| Orphan | □□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□□□□ | 177777777760700 | 1 (0.2) | 100 | 0 (0) | T1 |
| Orphan | ■□■■■■■■■□■■■■■■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | 577377777763771 | 1 (0.2) | 100 | 0 (0) | Manu2 |
| Orphan | □□■■□□□■□□■■□□■■■□□■■■■■■■■■■■□□□□■■■■■■■■■ | 142316377703771 | 1 (0.2) | 100 | 0 (0) | Unknown |
| Orphan | □□□□□□■■■□■■■■■■■■■■■□□□■■■■■■□□□□■■■■■■■■■ | 007377707703771 | 1 (0.2) | 100 | 0 (0) | Unknown |
| Orphan | ■□■■■■■■■□□□□□□□□□□□□□□□□□□□□□□■□□■■■■■■■■■ | 577000000023771 | 1 (0.2) | 100 | 0 (0) | Manu2 |
| Orphan | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■■■□■■■■■ | 000000000017571 | 1 (0.2) | 100 | 0 (0) | Unknown |
| Orphan | ■■■■■■■■■■■■□□□□□□□■■□□□□□□□■■■■□□■■■■■■■■■ | 777700300363771 | 1 (0.2) | 100 | 0 (0) | Manu2 |
| Orphan | □□■■■■■■■■■■□□□□□□□■■■■■□□□□□□□□□□■■■■■■■■■ | 177700370003771 | 1 (0.2) | 100 | 0 (0) | Unknown |
| Orphan | □□■■■■■■■■■■■■□■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | 177767777763771 | 1 (0.2) | 100 | 0 (0) | Manu2 |
| Orphan | ■□■■■■■■■■■■■■■■■■■■■■■■■■□□■■■■□□■■■■■■■■■ | 577777776363771 | 1 (0.2) | 100 | 0 (0) | Manu2 |
| Orphan | ■□■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■□□□□□■ | 577777777760401 | 1 (0.2) | 100 | 0 (0) | T1 |
| Orphan | ■□□□□□□□■■■■■■■■■■■■■■■■■■■■■■■■□□■■■■■■■■■ | 401777777763771 | 1 (0.2) | 100 | 0 (0) | Manu2 |
| Orphan | ■□■■■■■■■■■□■□□□□□□□□□□□■■■■■■■■□□■■■■■■■■■ | 577640007763771 | 1 (0.2) | 100 | 0 (0) | Manu2 |
SIT, spoligotype international shared type. A total of 58/61 SITs matched a preexisting shared type in the database, whereas 3/61 SITs were newly created either within the present study or after a match with an orphan in the database. A total of 22 SITs containing 438 isolates were clustered within this study (2 to 324 isolates per cluster), while 39 SIT plus 22 orphan (i.e., 61) isolates were unique. Note that SITs followed by an asterisk indicate a newly created shared type (n = 3 containing 4 isolates) due to ≥2 isolates belonging to an identical new pattern within this study or after a match with an orphan in the database. SIT 3309*—this study, n = 1; Colombia, n = 2; Ethiopia, n = 2. SIT 4011*—this study, n = 2. SIT 4012*—this study, n = 1; Brazil, n = 1.
Mixed infections and influencing factors.
Twelve (2.4%) of the 499 isolates were identified as mixed infections by 17 relevant loci of the standard 24-locus set, and the DST profiles of the 12 corresponding isolates are shown in Table 2. Using spoligotyping, isolates with mixed infections appeared in four lineages, including Manu2 (8/37 [21.62%]), Beijing (2/348 [0.57%]), T (1/83 [1.2%]), and unknown (1/18 [5.56%]). Eight of 12 (66.67%) isolates from patients with mixed infections belonged to the Manu2 lineage. Univariate logistic regression analysis showed that mixed infections were only significantly associated with Manu2 (OR, 47.72; P < 0.01; Table 3). Although there were no other bacterial or host variables (drug susceptibility, gender, age, or nationality) that were statistically significant (Table 3), a higher rate of mixed infection was shown in samples from patients of Han nationality (2.57%) and in drug-resistant isolates (2.65%) than those from minority patients (1.45%) and in susceptible isolates (1.22%), respectively. The 12 patients with mixed infections consisted of one young patient (0.85%), nine adults (2.8%), and two older patients (3.45%).
TABLE 2.
Genotyping and drug susceptibility test results of isolates from patients with mixed infections in the present study
| ID (no. of polymorphic alleles)a | DST resultb | Spoligotype | VNTR locus (no. of isolates with polymorphic alleles; HGDI)c |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T (5; 0.822) | U (4; 0.813) | F (2; 0.789) | O (5; 0.785) | H (3; 0.768) | N (3; 0.761) | I (2; 0.754) | V (2; 0.684) | C (5; 0.680) | D (2; 0.646) | J (6; 0.645) | G (1; 0.608) | K (3; 0.495) | Q (6; 0.408) | L (5; 0.260) | E (1; 0.221) | R (1; 0.113) | |||
| SC107 (2) | INHr, RIFr, STRr | Beijing | 4 | 5, 6 | 7, 8 | 5 | 4 | 4 | 3 | 0 | 3 | 2 | 3 | 3 | 4 | 4 | 2 | 5 | 3 |
| SC240 (2) | INHr, STRr | Manu2 | 4 | 8 | 6 | 5 | 4 | 1, 3 | 3 | 1 | 3 | 3 | 3 | 3 | 4 | 2, 4 | 2 | 5 | 3 |
| SSC0588 (2) | S | T1 | 0 | 9 | 4 | 1 | 3 | 3 | 2 | ND | 2 | 3 | 2 | 2 | 3, 4 | 2 | 2 | 5 | 3, 4 |
| SC081 (2) | INHr, RIFr, EMBr, STRr | Unknown | 6 | 6, 8 | 8 | 6 | 3, 5 | 2 | 2 | 1 | 2 | 2 | 3 | 2 | 4 | 4 | 2 | 5 | 3 |
| SSC0617 (3) | STRr | Beijing | 6 | 2 | 6 | 4 | 3 | 4 | 3 | ND | 3 | 3 | 2, 3 | 3 | 4 | 2, 4 | 2 | 4, 6 | 3 |
| SC023 (5) | INHr, STRr | Manu2 | 4, 5 | 7, 8 | 7 | 7 | 4 | 3, 6 | 3 | 2 | 1, 3 | 3 | 5 | 2 | 3 | 2 | 1, 2 | 5 | 3 |
| SSC0620 (5) | RIFr | Manu2 | 5, 6 | 5 | 7 | 4, 5 | 5 | 4 | 3 | 1, 2 | 3 | 2, 3 | 2, 4 | 2 | 4 | 4 | 2 | 5 | 3 |
| SSCM034 (5) | INHr, RIFr, STRr | Manu2 | 5 | ND | 4, 7 | 5, 7 | ND | 1 | 2, 3 | 2 | 2 | ND | 2, 3 | 2, 3 | 4 | 4 | 2 | ND | 3 |
| SSC0619 (7) | INHr | Manu2 | 1, 3 | 4, 8 | 2 | 3 | 3 | 2 | 3 | 2 | 2, 4 | 1, 4 | 2, 4 | 2 | 2 | 2, 5 | 1, 2 | 5 | 3 |
| SSC0407 (7) | INHr, RIFr, STRr | Manu2 | 3, 5 | 8 | 5 | 3, 6, 8 | 3 | 0 | 1 | 1 | 1, 2 | 2 | 3, 4 | 2 | 3, 4 | 1, 2 | 1, 2, 3 | 7 | 3 |
| SC262 (7) | INHr, RIFr, EMBr, STRr | Manu2 | 4, 6 | 8 | 5 | 3, 6 | 3, 5 | 2 | 2 | 1 | 1, 3 | 4 | 3 | 2 | 3, 4 | 2, 5 | 1, 2 | 5 | 3 |
| SC187 (9) | RIFr, EMBr | Manu2 | 5 | 7 | 4 | 2, 3 | 2, 5 | 1, 3 | 1, 2, 4 | 1, 3 | 1, 2, 3 | 2 | 3, 5 | 2 | 1 | 2, 4 | 1, 2, 3 | 5 | 3 |
ID, patient number.
DST, drug susceptibility test; RIFr, resistant to rifampin; INHr, resistant to isoniazid; STRr, resistant to streptomycin; S, pansusceptible; EMBr, resistant to ethambutol.
VNTR loci: C = 960, D = 1644, E = 2531, F = 2996, G = 3007, H = 3192, I = 4348, J = 802, K = 2165, L = 2461, N = 424, O = 1955, Q = 2401, R = 3171, T = 2163b, U = 4052, V = 4156. HGDI, Hunter-Gaston discriminatory index (the order of the VNTR locus was listed according to the HGDI from high to low); ND, not determined.
TABLE 3.
Potential factors associated with mixed infections by the univariate logistic analysisa
| Variable | Total no. of cases | No. of mixed infections | % of mixed infections | Univariate analysisb |
||
|---|---|---|---|---|---|---|
| OR | 95% CI | P value | ||||
| Patient | ||||||
| Gender | ||||||
| Male | 327 | 8 | 2.45 | 1.04 | 0.31–3.51 | 0.949 |
| Female | 170 | 4 | 2.35 | |||
| Nationality | ||||||
| Han | 428 | 11 | 2.57 | |||
| Minority | 69 | 1 | 1.45 | 0.56 | 0.07–4.39 | 0.579 |
| Age (yr) | ||||||
| Young (≤25) | 117 | 1 | 0.85 | |||
| Adult (25–55) | 321 | 9 | 2.80 | 3.35 | 0.42–26.70 | 0.254 |
| Old (≥55) | 58 | 2 | 3.45 | 4.14 | 0.37–46.66 | 0.250 |
| Bacterial | ||||||
| Drug susceptibility | ||||||
| Pansusceptible | 82 | 1 | 1.22 | |||
| Resistantc | 415 | 11 | 2.65 | 2.14 | 0.27–16.81 | 0.469 |
| Spoligotype | ||||||
| Beijing | 348 | 2 | 0.57 | |||
| Manu2 | 37 | 8 | 21.62 | 47.72 | 9.68–235.23 | 0.000 |
| Others | 114 | 2 | 1.75 | 3.09 | 0.43–22.19 | 0.262 |
The analysis was performed on 496 isolates of the total 499, as the clinical information for two patients was lost, and the age of one patient was unknown.
OR, odds ratio; 95% CI, 95% confidence interval.
Resistant to one or more drugs.
Mutation analysis and heteroresistance.
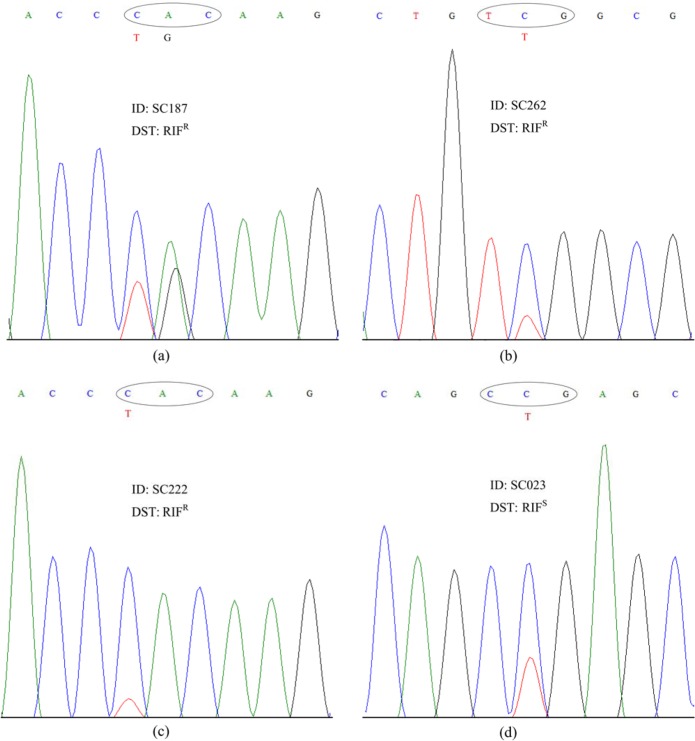
The randomly selected 292 isolates (78 RIF susceptible and 212 RIF resistant) from 499 isolates were sequenced on their rpoB gene, of which 218 isolates (74.66%) had mutations in their amplified region (see Table S1 in the supplemental material). When the phenotypic DST results were compared against the results of rpoB gene sequencing, 20 results were discordant (Table 4). Twelve (5.5%) of the 218 isolates were RIF susceptible, including two isolates of mixed infections (M. tuberculosis SC023 and SSC0588), and eight (10.81%) of the other 74 isolates with no mutation detected were RIF resistant, including four isolates of mixed infections (M. tuberculosis SSCM034, SC262, SSC0407, and SC187). Among the six isolates with mixed infections as described above, which had discordant results between their DST and mutation analysis, three were RIF heteroresistant and belonged to the Manu2 lineage, and their corresponding patients were infected with SC187, SC262, and SC023. Another RIF-heteroresistant isolate from a patient (SC222) had evidence of clonal heterogeneity (data not shown). In total, there were four isolates (1.37%) confirmed to be RIF heteroresistant, which had coexisting wild-type (WT) segments and mutations in the rpoB gene region, according to the sequencing results (Fig. 1).
TABLE 4.
Isolates identified as mixed infections by phenotypic drug susceptibility test for rifampin versus mutation analysis in rpoB genea
| Mutation category (no. of isolates) | No. (%) of isolatesb |
ID of mixed infections (ID with secondary peak) |
||
|---|---|---|---|---|
| RIFr | RIFs | RIFr | RIFs | |
| None (74) | 8 (10.81) | 66 (89.19) | SSCM034, SC262, SSC0407, SC187 | SSC0617, SSC0619, SC240, (SC023) |
| Single (177) | 165 (93.22) | 10 (5.65) | SC107, SSC0620, (SC187, SC262) | SSC0588, SC023 |
| Multiple (41) | 39 (95.12) | 2 (4.88) | SC081 | |
A total of 292 isolates that were sequenced on the rpoB gene were included in this statistical analysis. Two isolates for which clinical information was lost had a single mutation. The isolates of patients whose IDs are shown in parentheses were RIF heteroresistant.
RIFr, resistant to RIF; RIFs, susceptible to RIF.
FIG 1.
The four RIF heteroresistant isolates detected by DNA sequencing showed dual peaks of wild type (WT) and mutant bases at certain codons (circled). The phenotypic DST for RIF: RIFr (resistant to RIF) and RIFs (susceptible to RIF). (a to c) WT and secondary mutation bands at codons 526 (TGC), 531 (TTG), and 526 (TAC), respectively. (d) Main peak with mutant base (CCG) and WT (CTG) at codon 511.
DISCUSSION
The rates of mixed infections were first shown to be 3.4 to 14.1% by phage typing (1, 22) and were later confirmed to be 0.4 to 15.4% using DNA fingerprint analysis (23, 24) or PCR-based approaches in which primers were specific at the level of lineage only (3, 10). The rates of mixed infections ranged from 2.1 to 57.9% by 7 to 24 loci in MIRU-VNTR-based analysis in different regions (Table 5), and the rates of clonal heterogeneity were 1.3 to 9.3% (9, 25–27). One may speculate that the differences in the proportions of mixed infections are caused not only by differences in the methodology of detection, but they also might vary by geographic region. According to the available literature, there was no link between the detection rate of mixed infections and the discriminatory power of the loci (Table 5). Recently, 12% of mixed M. tuberculosis infections were identified by whole-genome sequencing (28); however, this method is limited by the cost, complexity of data analysis, and the short sequencing reads used to account for repeat regions (5). Mixed Beijing and non-Beijing strains caused the most pathogen-pathogen compatibility (10, 14, 29, 30); however, the mixed non-Beijing strains were also reported to be primary in some regions (12, 31). Besides, although Beijing strains were commonly present in mixed infections, their spoligotypes usually were masked by the spoligotypes of non-Beijing strains (14). Therefore, there might be some specific properties of the non-Beijing lineage that allow the stable coexistence of mixed strains of M. tuberculosis in a single host.
TABLE 5.
Mixed M. tuberculosis infections revealed by MIRU-VNTR-based methods across the literature
| Geographic regiona | Total no. of cases | No. (%) of mixed infectionsb | Corresponding SIT, lineage (n, %) of mixed infections | No. of loci (order of relevant loci by the no. of mixed infections detected)c | Reference or source |
|---|---|---|---|---|---|
| AFR | |||||
| Rwandad | 22 | 1 (4.3) | SIT52, T2 (1, 100) | 12 (J = H) | 43 |
| South Africae | 54 | 10 (18.5) | SIT53, T1 (10, 100) | 15 (T = O > C > V > M = Q = S > K = F > J = D = H > N) | 9 |
| Uganda (Kampala) | 113 | 8 (7.1) | No description | 15 (J > F > Q > D = H = M = O > T > K = C = B > S = U = V) | 8 |
| Malawi (Karonga) | 72 | 2 (2.8)f | LAM11_ZWE (1, 50); T1 (1, 50) | 24 (no report) | 31 |
| South Africa (KwaZulu-Natal) | 56 | 5 (9) | SIT34, S (1, 20); SIT52, T2 (1, 20); SIT54, Manu2 (1, 20); SIT1196, undesignated (1, 20); no SIT (1, 20) | 24 (M > J = K = E = L > F = C = H = N = Q = S = O = I = G = P = R) | 7 |
| Uganda (Mubende) | 72 | 8 (11.1) | SIT53/137,g T1/X2g (1, 12.5); SIT52, T2-Uganda I (1, 12.5); SIT420, T2-Uganda II (1, 12.5); others not reported | 15 (T > U > K = C = F > D = J > V = S = Q = N > B = H > M) | 46 |
| Botswana | 370 | 18 (4.9) | No description | 24 (no report) | 44 |
| EUR | |||||
| Belgium (Brussels) | 2 | 1 | No description | 12 (A = C = H = I) | 47 |
| Spain (Almería) | 780 | 11 (1.4) | No description | 24 (not clear) | 27 |
| MECA | |||||
| Bangladesh (Mymensingh) | 97 | 2 (2.1)f | Non-Beijing (2, 100) | 13 (H = B > M = J = D = F = S = N = C = V = X) | 15 |
| Georgia (near Tbilisi)h | 199 | 26 (13.1)f | No description | 15 (N > Q > C = F = H > M = J = D = K = U > V > S > O > T > B) | 2 |
| Kyrgyzstan (near Bishkek) | 56 | 3 (5.4) | SIT264, T5-RUS1 (1, 33.3); SIT53, T1 (1, 33.3); one not reported | 12 (F > A = C = H = I > D) | 30 |
| Uzbekistan (Tashkent)h | 7 | 4 (57.9) | SIT53, T1 (1, 25); SIT262, H3 (1, 25); SIT1196, undesignated (1, 25); unknown (1, 25) | 24 (T > J = O = K = Q = F > D = H = S > N = M = A = C = I) | 12 |
| FEA | |||||
| China (Shanghai)i | 249 | 2 (0.8) | Non-Beijing (1, 50); non-Beijing and Beijing (1, 50) | 7 (O > U = W) | 25 |
| Vietnam (Rural South)h | 1,248 | 60 (4.8) | EAI (33, 55); Manu2 (2, 3.3); Beijing (2, 3.3); CAS1-DELHI (1, 1.7); non-Beijing (1, 1.7); others not reported | 15 (no report) | 14 |
| China (Sichuan) | 499 | 12 (2.4) | SIT54, Manu2 (4, 33.3); SIT1096, Manu2 (3, 25); SIT1, Beijing (2, 16.7); orphan, Manu2 (1, 8.3); SIT2276, unknown (1, 8.3); SIT393, T1 (1, 8.3) | 24 (J = Q > T = O = C = L > U > H = N = K > F = I = V = D > G = E = R) | This study |
AFR, Africa; EUR, Europe; MECA, Middle-East and central Asia; FEA, Far-East Asia.
The proportion of mixed infections in some studies may not be representative of the underlying population, since patients in special circumstances, such as those from a hospital or prison or with an age in a specific range, were selected by the corresponding researchers.
A = 154, B = 580, C = 960, D = 1644, E = 2531, F = 2996, G = 3007, H = 3192, I = 4348, J = 802, K = 2165, L = 2461, M = 577, N = 424, O = 1955, P = 2347, Q = 2401, R = 3171, S = 3690, T = 2163b, U = 4052, V = 4156, W = VNTR3820, X = QUB1982, and the bold capital loci indicate three alleles.
The patients in Rwanda were from four provinces, Kigali, Butare, Ruhengeri, and Rwamagana.
The patients were from eight of the nine provinces of South Africa, including Eastern Cape, Limpopo, North West, Free State, Mpumalanga, Gauteng, KwaZulu-Natal, and Western Cape.
Mixed infections were detected by spoligotyping, IS6110-restriction fragment length polymorphism (RFLP), or PCR amplification using different primers and confirmed with the MIRU-VNTR method.
Corresponding SIT/lineage; the SIT53/T1 and SIT137/X2 isolates were from sputum and lymph node samples, respectively.
The number of mixed infections was different between the IS6110-RFLP method and the MIRU-VNTR method in this publication, while in our study, it was in accordance with the definition of the MIRU-VNTR method mentioned in the introduction in the text.
The isolates belonged to Beijing/non-Beijing based on deletion-targeted multiplex-PCR (DTM-PCR) genotyping, not spoligotyping.
Similar to findings in previous studies (see Table 5), non-Beijing spoligotype strains were significantly associated with mixed infections to a greater extent than Beijing ones (6.62% versus 0.57%; χ2, 16.41; P < 0.01) in Sichuan. Moreover, the prevalence of the non-Beijing family (30.26%) in this region was much higher than that in three provinces neighboring Sichuan to the north, including Tibet (9.62%), Gansu (12.5%), and Shanxi (20%) (32). Further statistical analysis revealed that Manu2 was the most likely lineage related to mixed infections in our study, and this lineage was also related to the orphan pattern (χ2, 28.09; P < 0.01). These two links supported the recent hypothesis that the misassignments or some undefined patterns in the SITVIT2 database might primarily be caused by mixed infections (6). Manu2 was identified as the lineage with the highest probability of being the admixture of two individual patterns in Sichuan. The similar situation also appeared in Vietnam, where the most isolates of mixed infections were found to belong to the East African-Indian (EAI) lineage (14). Compared with Beijing strains, EAI strains showed not only less virulence but also a 100-fold lower replication level in lung infections (33). It is noteworthy that the Manu and EAI lineages are believed to be closely related or even derived from a latest common ancestor (34).
Although a small number of resistant strains might not be detected by molecular methods because of the overgrowth of sensitive strains in drug-free medium, a majority of resistant strains in cases of mixed infections can be detected in drug selection medium. Isolates with mixed infections or heteroresistance usually show a contradiction between their phenotypic DST data and mutation analysis results (4, 11). Heteroresistance is a reason for the change in drug susceptibility patterns, and it was described for INH, RIF, EMB, STR, and fluoroquinolones by DST (35), line probe assays (11), and simultaneous detection of WT and mutated sequences by using PCR-based techniques or sequencing (11, 36–39). In our study, a low heteroresistance pattern rate was observed in the rpoB gene (1.37%), and similar rates have also been found in other regions, such as 1.4% in Italy (40), 1.9% in Finland and Russia (41), and 1.9% in Pakistan (42); however, other studies have reported significantly higher rates, such as 14.3% in Uzbekistan (12) and 28.8% in India (11). This discrepancy was reported to be related to differences in sample collection, detection method, or the prevalence of drug-resistant TB (12). Previous investigations reported that heteroresistance is primarily caused by mixed M. tuberculosis infections (12); similarly, in our study, 75% of the RIF heteroresistant isolates came from patients with mixed M. tuberculosis infections, and all belonged to Manu2 lineage. Therefore, the rate of heteroresistance may also vary with the level of mixed infections in a particular region.
Until now, about 5% of RIF-resistant isolates have had no mutation detected in the rpoB gene (19). In our study, 3.8% of the RIF-resistant isolates without any mutation detected were significantly associated with mixed infections (χ2, 56.78; P < 0.01), which was possibly because the mutations of the RIF-resistant isolates with mixed infections might be missed by DNA sequencing (4). It was reported that mixed infections might also be an important mechanism underlying the change in drug susceptibility patterns through the presence or absence of antibiotic pressure, which determined the dominant growth of strains of mixed infections (3). As the previous study suggested that there may be other genes or mechanisms conferring RIF resistance in TB (19), our data identified mixed infections as a possible reason for RIF-resistant isolates with no mutation in the rpoB gene region. Furthermore, with twice-reliable DST, we had already confirmed an extensively drug-resistant TB (XDR-TB) isolate, which was a combination of two different resistant strains, according to the sequencing result (data not shown), and this supports the idea that mixed infections increase the difficulty of diagnosis and treatment.
Nationality was considered for the first time as a factor influencing the occurrence of mixed infections; however, it was not significantly associated with mixed infections in our study, an observation that is similar to that with many other previously reported factors (Table 3) (7, 8, 43). From our data, there was a trend that the rate of mixed infections increased with age (0.85% to 2.8% to 3.45% in the young, adult, and older groups, respectively; Table 3). Older people, who normally have a poorer immune response, and those with immunosuppressive conditions, such as HIV infection, were considered to be associated with mixed infections to a higher extent (8, 44). The sensitivity of the Xpert MTB/RIF assay for RIF resistance detection was reduced in the cases of mixed infections and in HIV-infected patients who may suffer mixed infections (4). Our data suggest that Manu2 is the group proposing the highest risk of mixed infection in Sichuan. Besides, Manu lineage strains were recently reported to be associated with HIV infection (45), and Manu and EAI lineages were shown to be closely related. In a mouse model study, mice infected with EAI strains showed better survival than those infected with Beijing strains (33), which was similar to the phenomenon that patients with mixed infections had less extensive pulmonary pathology and increased immunological tolerance compared with those with single infections (14). Thus, the unique properties of the Manu2 lineage need further investigations in order to clarify its relevance to mixed infections.
It is realized that the minimal clinical information taken from the clinical records could not provide a highly concrete conclusion regarding the possible link between the occurrence of mixed infections, patient HIV status, and/or history of TB treatment. Indeed, the frequencies of mixed infections and RIF heteroresistance might be higher than those reported in the present study, since not all mixed infections were accurately identified by the MIRU-VNTR method that was reported to detect two mixed strains at a ratio of 1:99 (26); also, only one sample was taken from each patient. Other limitations include a lack of clear distinction between clonal heterogeneity and mixed infections (5), and Sanger sequencing may reduce sensitivity in the cases of mixed infection and heteroresistance compared to that with deep sequencing (4, 36).
In conclusion, this is the first study to describe situations of mixed infection and heteroresistance in Sichuan. The RIF heteroresistance pattern in this region was primarily caused by mixed infections, and the Manu2 group was the predominant lineage in the cases of mixed infections and RIF heteroresistance, which were greatly capable of superinfecting, cooperating with, or affecting the host immune response to coinfection. This is regarded as a possible part of the mechanisms conferring RIF resistance to isolates without any mutation in the rpoB gene region. Based on the present study, further studies are needed to clarify pathogen-pathogen compatibility, explain the characteristics of special lineages, such as Manu2 (described in our study), and establish a fast and effective method to detect, treat, and control both mixed and single infections of M. tuberculosis.
Supplementary Material
ACKNOWLEDGMENT
We thank the National Nature Science Foundation of China (grant 31200985) for their financial support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03507-14.
REFERENCES
- 1.Mankiewicz E, Liivak M. 1975. Phage types of mycobacterium tuberculosis in cultures isolated from Eskimo patients. Am Rev Respir Dis 111:307–312. [DOI] [PubMed] [Google Scholar]
- 2.Shamputa IC, Jugheli L, Sadradze N, Willery E, Portaels F, Supply P, Rigouts L. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir Res 7:99. doi: 10.1186/1465-9921-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Rie A, Victor TC, Richardson M, Johnson R, van der Spuy GD, Murray EJ, Beyers N, Gey van Pittius NC, van Helden PD, Warren RM. 2005. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med 172:636–642. doi: 10.1164/rccm.200503-449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zetola NM, Shin SS, Tumedi KA, Moeti K, Ncube R, Nicol M, Collman RG, Klausner JD, Modongo C. 2014. Mixed Mycobacterium tuberculosis complex infections and false-negative results for rifampin resistance by GeneXpert MTB/RIF are associated with poor clinical outcomes. J Clin Microbiol 52:2422–2429. doi: 10.1128/JCM.02489-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen T, van Helden PD, Wilson D, Colijn C, McLaughlin MM, Abubakar I, Warren RM. 2012. Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin Microbiol Rev 25:708–719. doi: 10.1128/CMR.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazzarini LCO, Rosenfeld J, Huard RC, Hill V, Lapa e Silva JR, DeSalle R, Rastogi N, Ho JL. 2012. Mycobacterium tuberculosis spoligotypes that may derive from mixed strain infections are revealed by a novel computational approach. Infect Genet Evol 12:798–806. doi: 10.1016/j.meegid.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Cohen T, Wilson D, Wallengren K, Samuel EY, Murray M. 2010. Mixed-strain Mycobacterium tuberculosis infections among patients dying in a hospital in KwaZulu-Natal, South Africa. J Clin Microbiol 49:385–388. doi: 10.1128/JCM.01378-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickman KR, Nabyonga L, Kateete DP, Katabazi FA, Asiimwe BB, Mayanja HK, Okwera A, Whalen C, Joloba ML. 2010. Detection of multiple strains of Mycobacterium tuberculosis using MIRU-VNTR in patients with pulmonary tuberculosis in Kampala, Uganda. BMC Infect Dis 10:349. doi: 10.1186/1471-2334-10-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavrum R, Mphahlele M, Ovreås K, Muthivhi T, Fourie PB, Weyer K, Grewal HMS. 2009. High diversity of Mycobacterium tuberculosis genotypes in South Africa and preponderance of mixed infections among ST53 isolates. J Clin Microbiol 47:1848–1856. doi: 10.1128/JCM.02167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanekom M, Streicher EM, Van de Berg D, Cox H, McDermid C, Bosman M, Gey van Pittius NC, Victor TC, Kidd M, van Soolingen D, van Helden PD, Warren RM. 2013. Population structure of mixed Mycobacterium tuberculosis infection is strain genotype and culture medium dependent. PLoS One 8:e70178. doi: 10.1371/journal.pone.0070178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Balooni V, Sharma BK, Kapil V, Sachdeva KS, Singh S. 2014. High degree of multi-drug resistance and heteroresistance in pulmonary TB patients from Punjab state of India. Tuberculosis (Edinb) 94:73–80. doi: 10.1016/j.tube.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann-Thiel S, van Ingen J, Feldmann K, Turaev L, Uzakova GT, Murmusaeva G, van Soolingen D, Hoffmann H. 2008. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur Respir J 33:368–374. doi: 10.1183/09031936.00089808. [DOI] [PubMed] [Google Scholar]
- 13.Zheng C, Zhao Y, Zhu G, Li S, Sun H, Feng Q, Luo M, Wu F, Li X, Hill V, Rastogi N, Sun Q. 2014. Suitability of IS6110-RFLP and MIRU-VNTR for differentiating spoligotyped drug-resistant Mycobacterium tuberculosis clinical isolates from Sichuan in China. Biomed Res Int 2014:763204. doi: 10.1155/2014/763204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huyen MNT, Kremer K, Lan NTN, Cobelens FGJ, Buu TN, Dung NH, Caws M, Tiemersma EW, van Soolingen D. 2012. Mixed tuberculosis infections in rural South Vietnam. J Clin Microbiol 50:1586–1592. doi: 10.1128/JCM.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamputa IC, Rigouts L, Eyongeta LA, El Aila NA, van Deun A, Salim AH, Willery E, Locht C, Supply P, Portaels F. 2004. Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J Clin Microbiol 42:5528–5536. doi: 10.1128/JCM.42.12.5528-5536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 29:2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang K, Sun H, Zhao Y, Guo J, Zhang C, Feng Q, He Y, Luo M, Li Y, Sun Q. 2013. Characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Sichuan in China. Tuberculosis (Edinb) 93:89–95. doi: 10.1016/j.tube.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demay C, Liens B, Burguiere T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N. 2012. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol 12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Bates JH, Stead WW, Rado TA. 1976. Phage type of tubercle bacilli isolated from patients with two or more sites of organ involvement. Am Rev Respir Dis 114:353–358. [DOI] [PubMed] [Google Scholar]
- 23.Das S, Narayanan S, Hari L, Mohan NS, Somasundaram S, Selvakumar N, Narayanan PR. 2004. Simultaneous infection with multiple strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int J Tuberc Lung Dis 8:267–270. [PubMed] [Google Scholar]
- 24.du Plessis DG, Warren R, Richardson M, Joubert JJ, van Helden PD. 2001. Demonstration of reinfection and reactivation in HIV-negative autopsied cases of secondary tuberculosis: multilesional genotyping of Mycobacterium tuberculosis utilizing IS 6110 and other repetitive element-based DNA fingerprinting. Tuberculosis (Edinb) 81:211–220. doi: 10.1054/tube.2000.0278. [DOI] [PubMed] [Google Scholar]
- 25.Fang R, Li X, Li J, Wu J, Shen X, Gui X, DeRiemer K, Liu L, Mei J, Gao Q. 2008. Mixed infections of Mycobacterium tuberculosis in tuberculosis patients in Shanghai, China. Tuberculosis 88:469–473. doi: 10.1016/j.tube.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia de Viedma D, Alonso Rodriguez N, Andres S, Ruiz Serrano MJ, Bouza E. 2005. Characterization of clonal complexity in tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat typing. J Clin Microbiol 43:5660–5664. doi: 10.1128/JCM.43.11.5660-5664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro Y, Herranz M, Perez-Lago L, Martinez Lirola M, INDAL-TB, Ruiz-Serrano MJ, Bouza E, Garcia de Viedma D. 2011. Systematic survey of clonal complexity in tuberculosis at a populational level and detailed characterization of the isolates involved. J Clin Microbiol 49:4131–4137. doi: 10.1128/JCM.05203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant JM, Harris SR, Parkhill J, Dawson R, Diacon AH, van Helden P, Pym A, Mahayiddin AA, Chuchottaworn C, Sanne IM, Louw C, Boeree MJ, Hoelscher M, McHugh TD, Bateson AL, Hunt RD, Mwaigwisya S, Wright L, Gillespie SH, Bentley SD. 2013. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir Med 1:786–792. doi: 10.1016/S2213-2600(13)70231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox HS, Kubica T, Doshetov D, Kebede Y, Rusch-Gerdess S, Niemann S. 2005. The Beijing genotype and drug resistant tuberculosis in the Aral Sea region of central Asia. Respir Res 6:134. doi: 10.1186/1465-9921-6-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mokrousov I, Valcheva V, Sovhozova N, Aldashev A, Rastogi N, Isakova J. 2009. Penitentiary population of Mycobacterium tuberculosis in Kyrgyzstan: exceptionally high prevalence of the Beijing genotype and its Russia-specific subtype. Infect Genet Evol 9:1400–1405. doi: 10.1016/j.meegid.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Mallard K, McNerney R, Crampin AC, Houben R, Ndlovu R, Munthali L, Warren RM, French N, Glynn JR. 2010. Molecular detection of mixed infections of Mycobacterium tuberculosis strains in sputum samples from patients in Karonga District, Malawi. J Clin Microbiol 48:4512–4518. doi: 10.1128/JCM.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Feng Q, Tang K, Zhang C, Sun H, Luo T, Yang Z, Couvin D, Rastogi N, Sun Q. 2012. The population structure of drug-resistant Mycobacterium tuberculosis clinical isolates from Sichuan in China. Infect Genet Evol 12:718–724. doi: 10.1016/j.meegid.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 33.Reiling N, Homolka S, Walter K, Brandenburg J, Niwinski L, Ernst M, Herzmann C, Lange C, Diel R, Ehlers S, Niemann S. 2013. Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice. mBio 4:e00250-13. doi: 10.1128/mBio.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas SK, Iravatham CC, Moni BH, Kumar A, Archana BV, Majid M, Priyadarshini Y, Rani PS, Valluri V, Hasnain SE, Ahmed N. 2011. Modern and ancestral genotypes of Mycobacterium tuberculosis from Andhra Pradesh, India. PLoS One 6:e27584. doi: 10.1371/journal.pone.0027584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adjers-Koskela K, Katila ML. 2003. Susceptibility testing with the manual mycobacteria growth indicator tube (MGIT) and the MGIT 960 system provides rapid and reliable verification of multidrug-resistant tuberculosis. J Clin Microbiol 41:1235–1239. doi: 10.1128/JCM.41.3.1235-1239.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eilertson B, Maruri F, Blackman A, Herrera M, Samuels DC, Sterling TR. 2014. High proportion of heteroresistance in gyrA and gyrB in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 58:3270–3275. doi: 10.1128/AAC.02066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karahan ZC, Akar N. 2005. Restriction endonuclease analysis as a solution for determining rifampin resistance mutations by automated DNA sequencing in heteroresistant Mycobacterium tuberculosis strains. Microb Drug Resist 11:137–140. doi: 10.1089/mdr.2005.11.137. [DOI] [PubMed] [Google Scholar]
- 38.Lau RW, Ho PL, Kao RY, Siu GK, Cheng VC, Yuen KY, Yam WC. 2010. Rapid diagnosis of multidrug-resistant smear-positive pulmonary tuberculosis. Int J Antimicrob Agents 35:202–203. doi: 10.1016/j.ijantimicag.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 39.Rinder H, Mieskes KT, Loscher T. 2001. Heteroresistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis 5:339–345. [PubMed] [Google Scholar]
- 40.Miotto P, Piana F, Penati V, Canducci F, Migliori GB, Cirillo DM. 2006. Use of genotype MTBDR assay for molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical strains isolated in Italy. J Clin Microbiol 44:2485–2491. doi: 10.1128/JCM.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mäkinen J, Marttila HJ, Marjamäki M, Viljanen MK, Soini H. 2006. Comparison of two commercially available DNA line probe assays for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol 44:350–352. doi: 10.1128/JCM.44.2.350-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farooqi JQ, Khan E, Alam SM, Ali A, Hasan Z, Hasan R. 2012. Line probe assay for detection of rifampicin and isoniazid resistant tuberculosis in Pakistan. J Pak Med Assoc 62:767–772. [PubMed] [Google Scholar]
- 43.Umubyeyi AN, Shamputa IC, Rigouts L, Dediste A, Karita E, Struelens MJ, Portaels F. 2007. Molecular investigation of recurrent tuberculosis in patients from Rwanda. Int J Tuberc Lung Dis 11:860–867. [PubMed] [Google Scholar]
- 44.Shin SS, Modongo C, Ncube R, Sepako E, Klausner JD, Zetola NM. 2014. Advanced immune suppression is associated with increased prevalence of mixed-strain Mycobacterium tuberculosis infections among persons at high risk for drug-resistant tuberculosis in Botswana. J Infect Dis 11:347–351. doi: 10.1093/infdis/jiu421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belay M, Ameni G, Bjune G, Couvin D, Rastogi N, Abebe F. 2014. Strain diversity of Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients in Afar pastoral region of Ethiopia. Biomed Res Int 2014:238532. doi: 10.1155/2014/238532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muwonge A, Kankya C, Olea-Popelka F, Biffa D, Ssengooba W, Berit D, Skjerve E, Johansen TB. 2013. Molecular investigation of multiple strain infections in patients with tuberculosis in Mubende district, Uganda. Infect Genet Evol 17:16–22. doi: 10.1016/j.meegid.2013.03.039. [DOI] [PubMed] [Google Scholar]
- 47.Allix C, Supply P, Fauville-Dufaux M. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin Infect Dis 39:783–789. doi: 10.1086/423383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.