FIG 3.
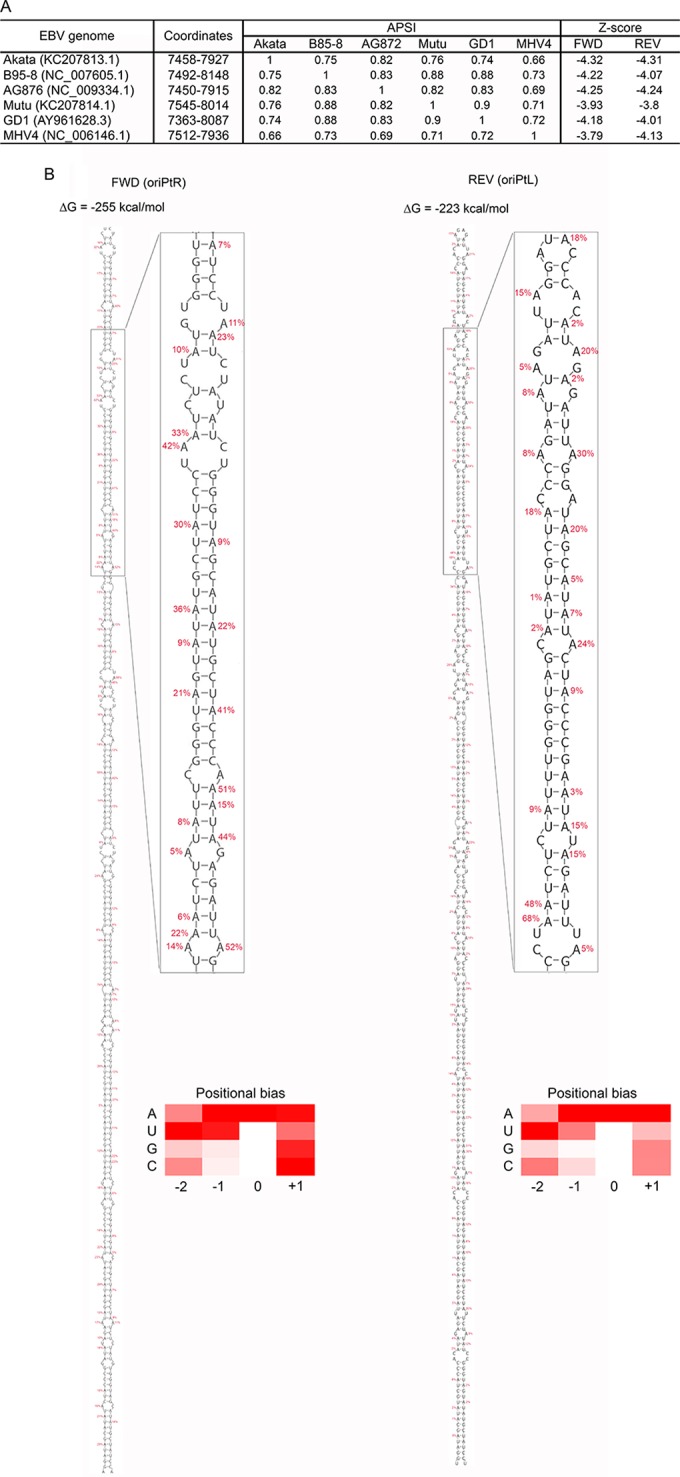
oriPt secondary structures. (A) Consensus hairpin structures were predicted in the FR region of oriP across five EBV strains (Akata, B95-8, G876, Mutu, and GD1) and macacine herpesvirus 4 (MHV4). The average pairwise sequence identity (APSI) was calculated for the regions where the hairpin structures were identified in the six virus strains. The z-scores were calculated for the hairpin structures in sense (Fwd) and antisense (Rev) transcripts in each virus. (B) Predicted hairpin structures for oriPtR (left) and oriPtL (right) with associated folding free energy values. Editing frequencies for each adenosine through each hairpin are shown (an expanded view is depicted in Fig. S3 in the supplemental material for visualization in greater detail). Heat maps show relative average editing for A's, U's, G's, and C's located at positions −2, −1, 0, and +1 relative to the edited adenosine residue (0 position).
