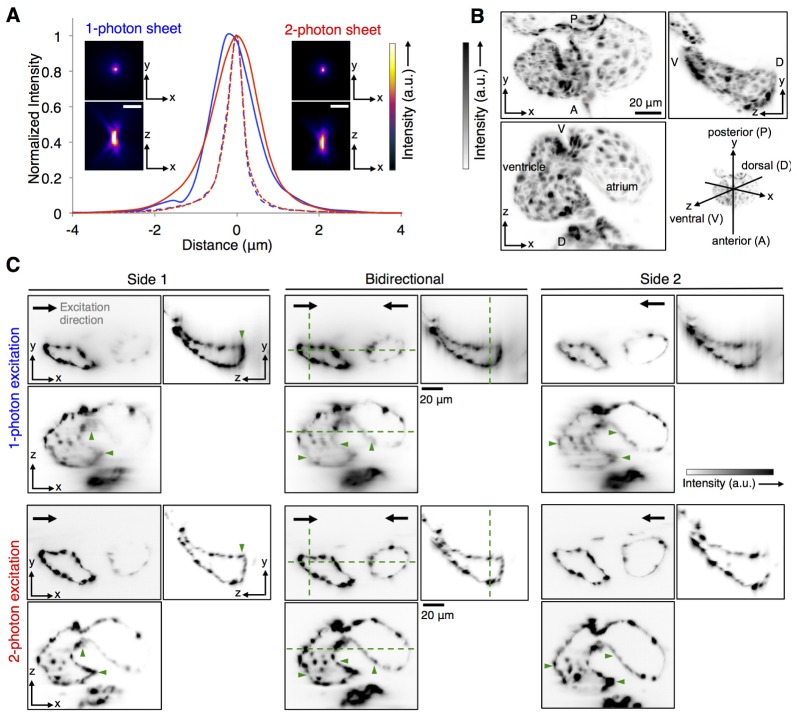
Fig. 3.
Characterization of imaging performance of 1p-SPIM and 2p-SPIM. (A) Resolution estimates from imaging 170nm-diameter fluorescent beads embedded in agarose. Images show the maximum intensity projection (MIP) along the xy and xz planes, for the same bead imaged by the two modalities. Plot shows the signal line profiles along the axial z-axis (solid line) and lateral x-axis (dashed line) over the bead images, showing FWHM axial resolution of ~1.5 µm and lateral resolution of ~0.5 µm. (B, C) Comparison of penetration depth of 1p-SPIM and 2p-SPIM in imaging the heart of a 60-hpf transgenic zebrafish, Tg(kdrl:GFP) s843, where the endocardium cell layer is labeled with GFP. The heart beating was stopped with a high concentration of anesthetic, allowing conventional z-stack imaging to capture the 3D structure. (B) MIPs of the 2p-SPIM data set along the three principal Cartesian planes are shown to aid the visualization of the anatomy of the heart. (C) Orthogonal views of the 3D data sets, showing single image slices at particular planes (designated by dashed green lines in the central image panels), to demonstrate the penetration depth of 1p-SPIM and 2p-SPIM. 3D images were taken with either uni-directional (light sheet coming from side 1 or side 2) or bidirectional (light sheets coming from both sides simultaneously) excitation. To facilitate fair comparison between the two imaging modalities, best effort was made to show the same planes within the heart for each data set (with the constraint that the live sample did shift slightly between data sets), with linear display contrast minimum and maximum set equal to the minimum and maximum pixel value, respectively, in each of the presented image slice. Overall, we see that 1p-SPIM and 2p-SPIM performs equally well in resolving the endocardium wall closer to the outer surface of the embryo, but 2p-SPIM performs better at capturing cardiac structures deeper inside the animal, as highlighted by structures marked with green arrowheads.