Fig. 3.
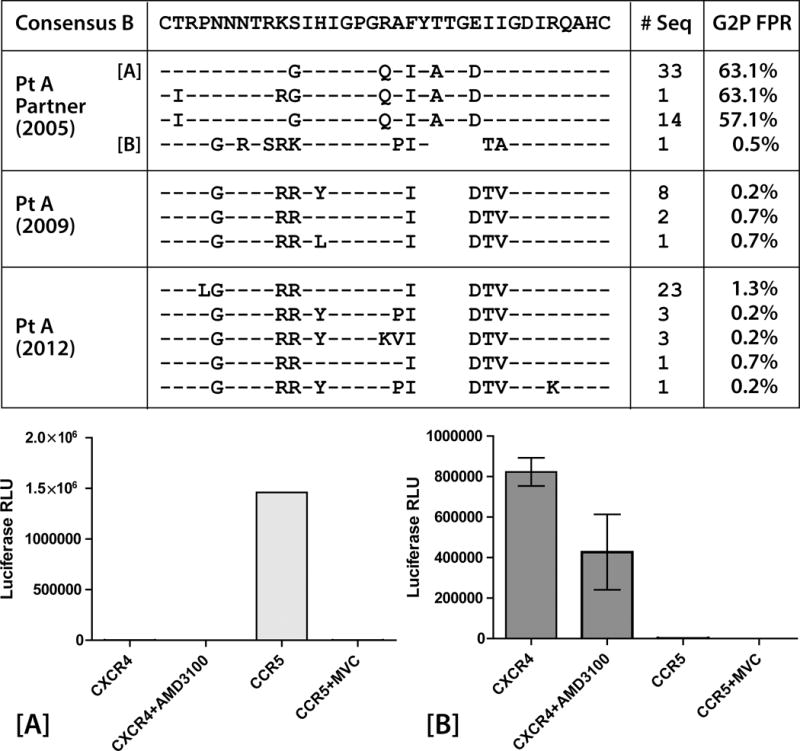
Conservation alignment of HIV-1 env V3 loop amino acid sequences from single genome PCR amplification and sequencing from participant A before and after virologic escape (2009 and 2012 respectively) and his sexual partner in 2005. One V3 loop sequence from participant A’s partner was more similar to V3 loop sequences from participant A, with a GPGR V3 crown motif in lieu of GPGQ. A positively charged amino acid in V3 loop position 11 (arginine) in all of participant A’s sequences and the related V3 loop sequence from his partner (lysine) was observed, and the geno2pheno (G2P) algorithm predict X4-D/M virus (FPR = false positive rate of classifying an R5-virus as X4) from these sequences. Results from phenotypic coreceptor usage assays using pseudoviruses incorporating single genome envelope sequences from participants A’s partner are shown in the bottom frame. Pseudovirus incorporating a representative sequence from participantA partner’s majority V3 loop sequences [A] used only CCR5 for entry. Virus incorporating the partner envelope sequence predicted to be X4-D/M [B] used only CXCR4. Error bars represent standard error measurement from triplicate wells of each phenotypic assay.
