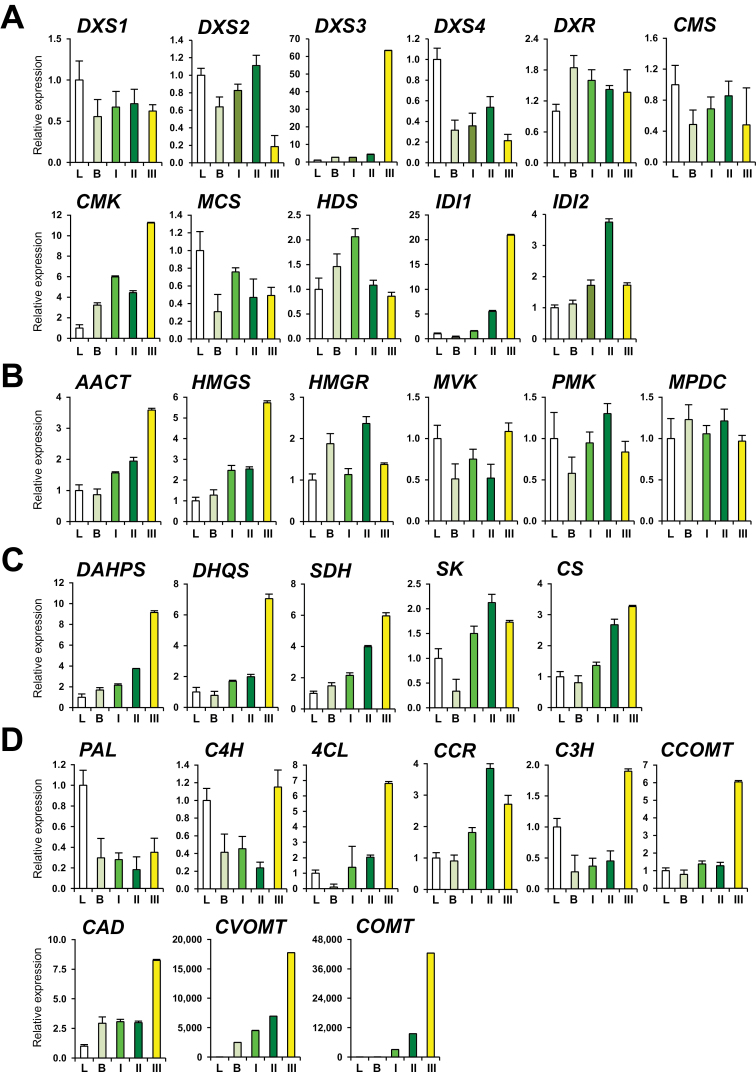
Fig. 3.
qRT-PCR analyses of different biosynthetic pathway genes. Expression of genes involved in the MEP (A), MVA (B), shikimate (C), and phenyl propanoid (D) biosynthetic pathways were examined from leaves (L), buds (B), and three different stages of flower development: undeveloped small flowers (I), mature green flowers (II), and fully mature yellow flowers (III) by qRT- PCR. DXS, 1-deoxy-d-xylulose 5- phosphate synthase; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; CMS, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase; MCS, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; HDS, 4- hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; IDI, isopentenyl pyrophosphate isomerase; AACT, acetyl-CoA acetyltransferase; HMGS, hydroxymethylglutaryl-CoA synthase; HMGR, hydroxymethylglutaryl-CoA reductase; MVK, mevalonate kinase; PMK, phosphomevalonate kinase; MPDC, mevalonate diphosphate decarboxylase; DAHPS, 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase; DHQS, 3-dehydroquinate synthase; SDH, shikimate dehydrogenase; SK, shikimate kinase; CS, chorismate synthase; PAL, phenylalanine ammonia lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumaroyl-CoA ligase; CCR, cinnamoyl-CoA reductase; C3H, p-coumarate-3-hydroxylase; CCOMT, caffeoyl-CoA 3-O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; CVOMT, chavicol O-methyltransferase; COMT, caffeic acid/5-hydroxyferulic acid O-methyltransferase. (This figure is available in colour at JXB online.)