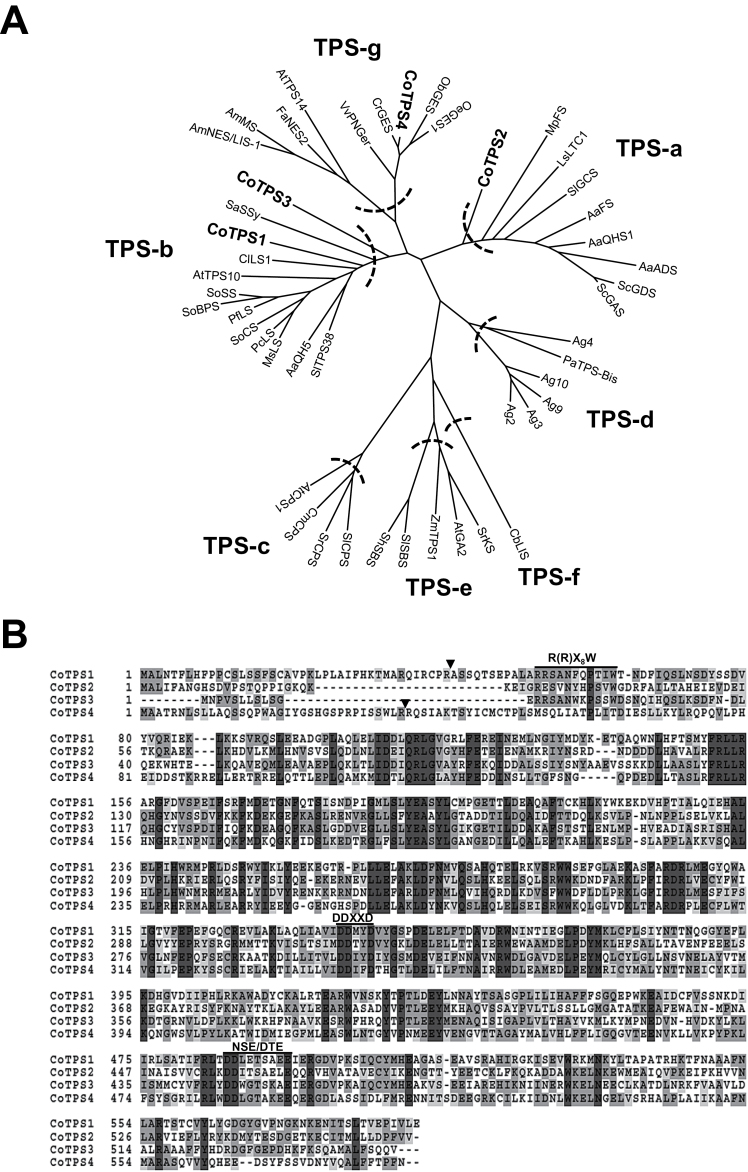
Fig. 4.
Phylogenetic analysis and alignment of TPSs from dwarf ylang ylang. (A) A maximum likelihood tree was drawn using the MEGA 6 program from an alignment of full-length CoTPSs with other plant TPSs. The seven TPS subfamilies a–g are delimited by dashed lines, based on the taxonomic distribution of the TPS families (Chen et al., 2011). AaADS, Artemisia annua (Aa) amorpha-4,11-diene synthase (NCBI Protein no. AFA34434); AaFS, β-farnesene synthase (Q9FXY7); AaQHS1, β-caryophyllene synthase (AAL79181); AaQH5, linalool synthase (AAF13356); Ag10, Abies grandis (Ag) 4S-limonene synthase (AAB70907); Ag2, myrcene synthase (AAB71084); Ag4, δ-selinene synthase (AAC05727); Ag3, pinene synthase (AAB71085); Ag9, terpinolene synthase (AAF61454); AmNES/LIS-1, Antirrhium majus (Am) nerolidol/linalool synthase1 (ABR24417); AmMS, myrcene synthase (AAO41727); AtCPS1, Arabidopsis thaliana (At) copalyl diphosphate synthase (NP_192187); AtGA2, kaurene synthase; AtTPS10 (AAC39443), myrcene/ocimene synthase (AAG09310); AtTPS14, linalool synthase (NP176361); CbLIS, Clarkia breweri S-linalool synthase (AAC49395); ClLS1, Citrus limon limonene synthase 1 (AAM53944); CmCPS, Cucurbits maxima copalyl diphosphate synthase (AAD04292); CrGES, Catharanthus roseus geraniol synthase (AFD64744); FaNES2, Fragaria×ananassa nerolidol synthase (CAD57081); LsLTC1, Lactuca sativus germacrene A synthase (AAM11626); MpFS, Mentha piperita β-farnesene synthase (AAB95209); MsLS, Mentha spicata 4S-limonene synthase (AAC37366); ObGES, Ocimum basilicum geraniol synthase (AAR11765); OeGES1, Olea europaea geraniol synthase 1 (AFI47926); PaTPS-Bis, Picea abies α-bisabolene synthase (AAS47689); PcLS, Perilla citriodora limonene synthase (AAG31435); PfLS, Perilla frutescens linalool synthase (AAL38029); SaSSy, Santalum album santalene/bergamotene synthase (ADO87000); ScGAS, Solidago canadensis (Sc) germacrene A synthase (CAC36896); ScGDS, germacrene D synthase (AAR31145); ShSBS, Solanum habrochaites santalene/bergamotene synthase (B8XA41); SlSBS, Solanum lycopersicum (Sl) santalene and bergamotene synthase (XP004244438); SlCPS, copalyl diphosphate synthase (BAA84918); SlGCS, germacrene C synthase (AAC39432); santalene/bergamotene synthase (BAA84918); Sl TPS38, Sl terpene synthase 38 (AEP82768); SoBPS, Salvia officinalis (So) bornyl diphosphate synthase (AAC26017); SoCS, 1,8-cineole synthase (AAC26016); SoSS, sabinene synthase (AAC26018); SrCPS, Stevia rebaudiana (Sr) copalyl pyrophosphate synthase (AAB87091); SrKS, kaurene synthase (AAD34294); VvPNGer, Vitis vinifera geraniol synthase (ADR74218); ZmTPS1, Zea mays terpene synthase 1 (AAO18435). (B) Comparison of deduced amino acid sequences of dwarf ylang ylang TPSs. The deduced amino acid sequences of CoTPS genes were aligned using CLUSTAL W. The Asp-rich domain DDXXD, the R(R)X8W motif, and the NSE/DTE motif, which are highly conserved in plant TPSs and required for TPS activity, are indicated. Arrowheads denote the predicted cleavage sites of plastidial transit peptides. Completely conserved residues are shaded in dark grey, identical residues in grey, and similar residues in light grey. Dashes indicate gaps introduced to maximize sequence alignment.