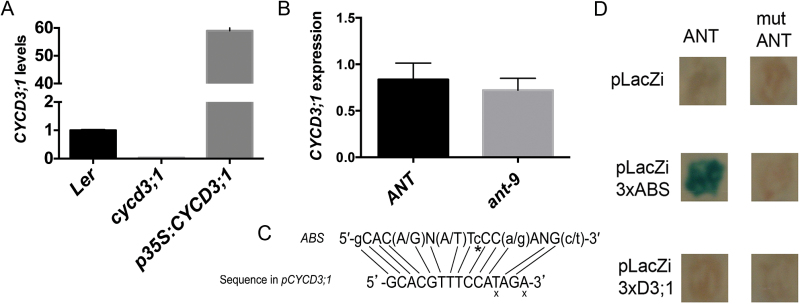
Fig. 4.
qPCR analysis of CYCD3;1 transcripts in WT Ler, cycd3;1, and p35S:CYCD3;1 shoots (A) and WT Ler and ant-9 mutant flowers (stage 1–12; Smyth et al., 1990) (B). In (A), error bars represent the SD from three technical replicates. In (B), error bars represent the SD from four biological replicates. Each replicate contained three inflorescences from an individual plant: the apical inflorescence, and the two youngest thereafter. WT transcript levels were set to 1.0 in both cases. (C) The sequence located 174bp upstream of the CYCD3;1 open reading frame that is similar to the ANT-binding site. An x indicates bases that do not match those at equivalent positions in the ABS. * indicates a base that is missing in the CYCD3;1 promoter sequence. (D) Yeast one-hybrid assay testing the binding of ANT to a putative ANT-binding site in the CYCD3;1 promoter. ANT (left) and dominant-negative ant (right) genes were expressed in pGAD424 lacking the GAL4 activation domain (Krizek, 2003). The pLacZi reporter vector was either empty (top), contained three copies of the optimal ANT-binding site (middle; Nole-Wilson and Krizek, 2000; Krizek, 2003), or contained three copies of the putative ANT-binding site in the CYCD3;1 promoter. These motifs were upstream of the TATA box of the yeast CYC1 gene fused to β-galactosidase in pLacZi. An X-gal assay was performed. Only WT ANT transactivates the downstream β-galactosidase reporter to detectable levels.