Fig. 5.
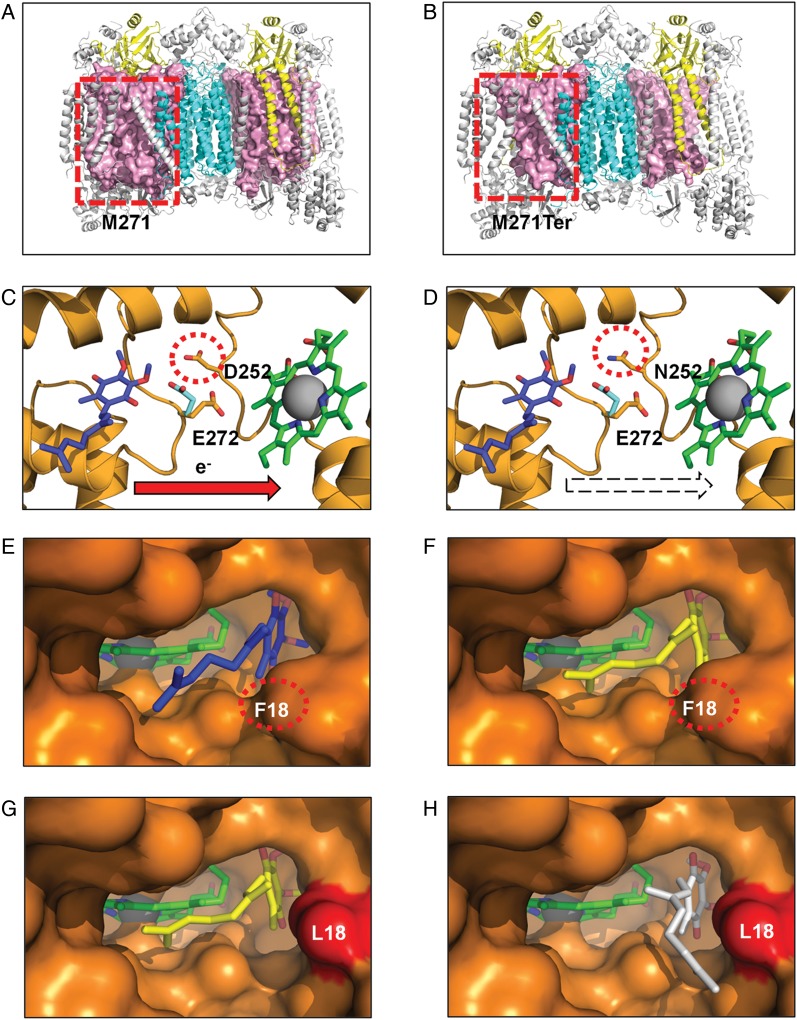
Structural consequences of 3 classes of functional mutation present in GBM complex III and IV mitochondrial proteins (colored as in Fig. 2). Frameshift mutation. (A) Wild-type (M271) residue in MT-CO1 is rendered as a space-filling model (light pink), highlighting its central position within each monomer. (B) The mutant A6692del (M271Ter) results in deletion of >47% of the MT-CO1 polypeptide chain, disrupting the interaction of multiple subunits that surround MT-CO1. Active site mutation. (C) Wild-type (D252) residue within the MT-CYB complex III Qo site. (D) The mutant (N252) may affect the passage of protons from the hydroxyquinone (blue) bound at the Qo site to the solvent phase. Residue E272 is shown in the 2 alternate conformations observed in x-ray crystal structures with the stigmatellin-inhibited complex (PDB ID 1KB950) (cyan) and in the HHDBT (5-n-heptyl-6-hydroxy-4, 7-dioxobenzothiazole)-inhibited complex (PDB ID 1P8432) (orange) and heme bL. The quinol binding site is inferred from the inhibitors binding site. Binding pocket mutation in complex III Qi site. (E) The wild-type Qi-site pocket is shown as a surface representation with the deeply buried heme group and bound ubiquinone (PDB ID 1NTZ34) (blue). (F) The results from in silico docking studies confirm that the predicted hydroxyquinone position (yellow) corresponds closely to that observed in the crystal structure. (G) An extra cavity is shown where the F18L GBM mutation (red) opens up the binding pocket. (H) The hydroxyquinone is observed to occupy additional binding sites (white) that are not possible in the wild-type protein due to steric constraints provided by the aromatic F18 residue. Since this highly conserved pocket is tailored precisely for hydroxyubiquinone binding, such a change is likely to interfere with its association/dissociation and ultimately efficient electron transfer from heme bH.