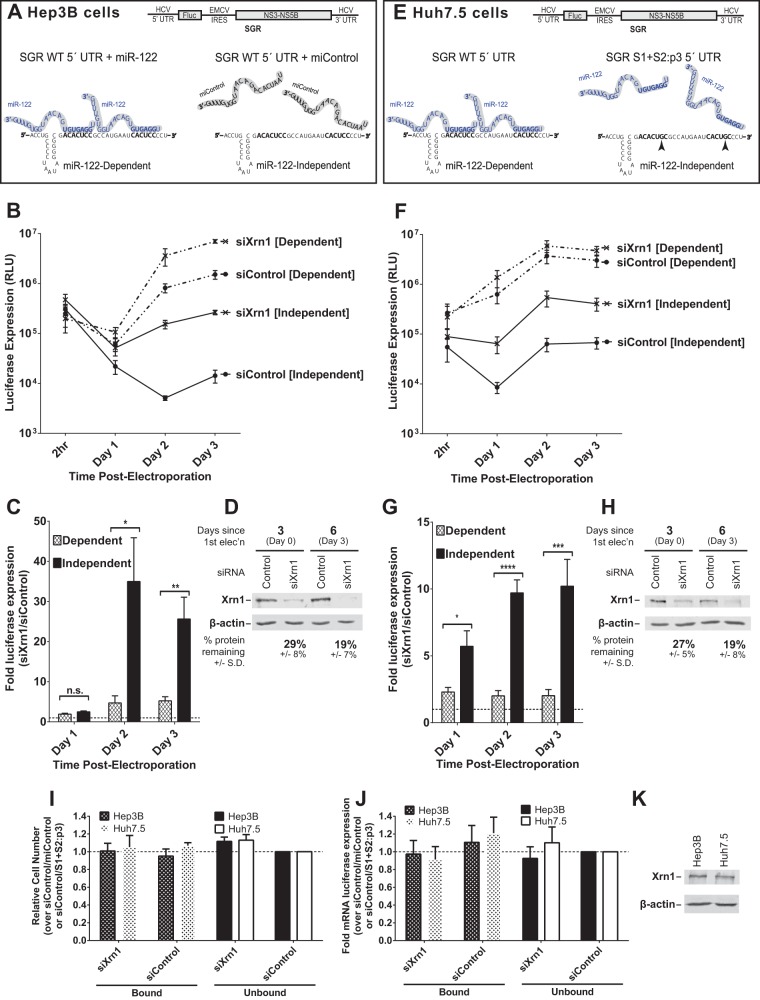
FIG 1.
Xrn1 knockdown increases miR-122-independent replication of subgenomic JFH-1 HCV RNA more than it increases miR-122-dependent replication. (A) Cartoon of experimental systems. Top, depiction of subgenomic JFH-1 HCV RNA (SGR), which contains a firefly luciferase reporter gene expressed from the HCV IRES and the viral nonstructural proteins expressed from an EMCV IRES; bottom, depiction of miR-122 binding sites on the 5′ UTR of HCV RNA. Hep3B cells lack expression of detectable miR-122 but can be supplemented with either synthetic miR-122 (left; binding at both sites, to achieve miR-122-dependent replication) or miControl (right; no binding, to achieve miR-122-independent replication) as shown. (B) Hep3B cells were electroporated with siXrn1 or siControl for preknockdown, and 3 days later (day 0) cells were electroporated again with the indicated siRNA, wild-type SGR HCV RNA, and miR-122 (miR-122 dependent) or miControl (miR-122 independent). Replication was measured by evaluating luciferase production at the indicated time points post-second electroporation. (C) The effect of Xrn1 knockdown on miR-122-dependent and miR-122-independent subgenomic HCV RNA replication in Hep3B cells was determined by measuring the fold increase in luciferase expression with siXrn1 treatment over luciferase expression with no knockdown (siControl), using luciferase data from panel B at the indicated time points. Significance was determined by paired parametric t test. (D) The effectiveness of siXrn1 at reducing Xrn1 protein levels in Hep3B cells was determined by Western blotting with antibodies against Xrn1 and β-actin. A representative blot is shown depicting Xrn1 protein levels 3 days post-first electroporation (at the time of second electroporation, which includes viral and miRNAs) in the first two lanes, and 6 days post-first electroporation or 3 days post-second electroporation. Percent knockdown ± standard deviation relative to the siControl-treated cells was determined by infrared fluorescence quantification on blots from three or more independent experiments. (E) Cartoon of experimental systems. Top, depiction of subgenomic JFH-1 HCV RNA (SGR); bottom, depiction of miR-122 binding at the 5′ UTR of HCV RNA. Huh7.5 cells endogenously express miR-122, and all wild-type replication is miR-122 dependent (left), so to study miR-122-independent replication, miR-122 binding was abolished by mutating both miR-122 binding sites (right). (F) Huh7.5 cells were treated as described in the legend to panel B, but no miRNA was added since Huh7.5 cells already express endogenous miR-122. “Dependent” samples were electroporated with wild-type SGR RNA, while “independent” samples were electroporated with the miR-122 binding site mutant SGR S1+S2:p3 RNA. (G) The fold increase in miR-122-dependent and -independent HCV replication induced by Xrn1 knockdown in Huh7.5 cells was determined as described in the legend to panel C. Significance was determined by unpaired parametric t test. (H) Xrn1 protein knockdown efficiency was determined by Western blot analysis of protein from three independent experiments as described in the legend to panel D, and a representative blot is shown. (I) The effect of siXrn1, viral RNAs, and microRNAs on cell survival was evaluated by WST-1 3 days postelectroporation from samples in panels B and F, and cell numbers are normalized to siControl-miControl or siControl-SGR S1+S2:p3 samples. (J) Transfection efficiency in the experiments in panels B and F was evaluated 2 h postelectroporation by measuring Renilla luciferase expression from a coelectroporated mRNA. Samples are normalized to siControl-miControl or siControl-SGR S1+S2:p3 samples. (K) Untreated Hep3B and Huh7.5 cell lysates were Western blotted to show steady-state levels of Xrn1 protein in each cell type.