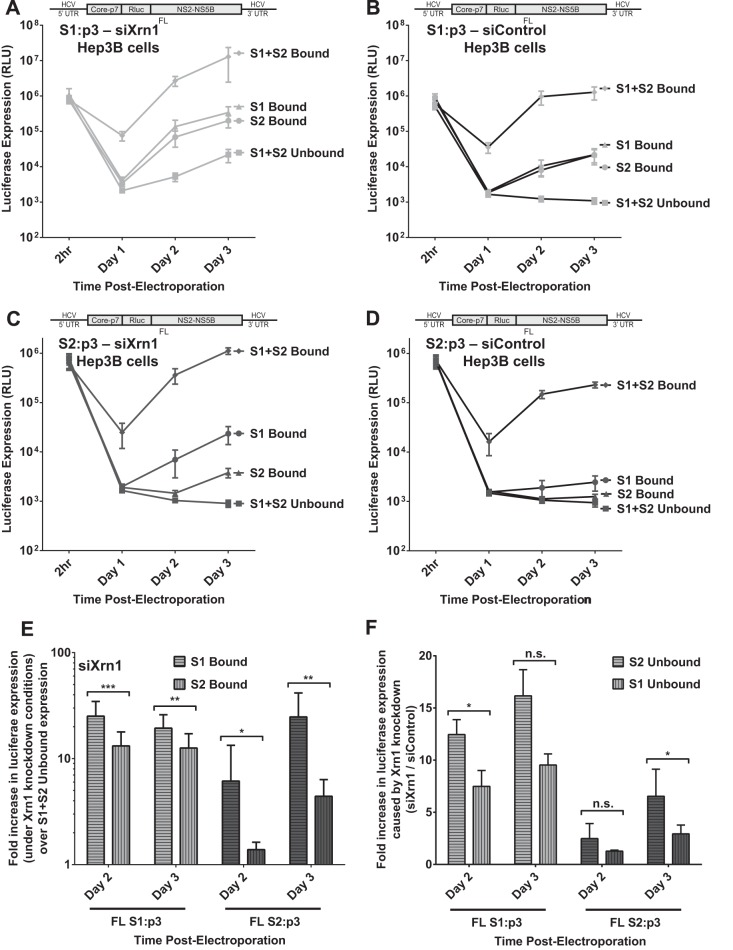
FIG 8.
The impact of miR-122 binding at site 1 compared to binding at site 2 with full-length HCV RNA when Xrn1 is knocked down. To assess the impact of miR-122 binding to site 1 or site 2 independent from that of protection from Xrn1, we tested the influence of miR-122 on replication of HCV in Xrn1-depleted cells. (A and B) We performed a time course of FL S1:p3 viral RNA replication in Hep3B cells with Xrn1 knockdown (A) or no knockdown (B). Replication of FL S1:p3 was assessed following supplementation with miR-122/p3 (S1 Bound), miR-122 (S2 Bound), miR-122/p3 and miR-122 (S1+S2 Bound), or miControl (S1+S2 Unbound) as depicted in Fig. 5Aiii, and replication was measured by evaluating reporter Renilla luciferase expression at the indicated time points. Results are an average from five independent experiments. (C and D) Xrn1 (C)- and siControl (D)-treated Hep3B cells were treated as described for panels A and B, respectively, but were electroporated with FL S2:p3 RNA, with miRNAs resulting in miR-122 binding site occupation as depicted in Fig. 5Aiv. Results are an average from five independent experiments. (E) In the context of Xrn1 knockdown, the fold increase in replication of FL S1:p3 RNA (light gray, left) and FL S2:p3 RNA (dark gray, right) when either S1 or S2 is bound was compared to unbound replication. Significance was determined using a ratio-paired parametric t test of relevant samples. (F) The effect of Xrn1 knockdown on FL S1:p3 or FL S2:p3 with either site 1 or site 2 unoccupied is shown by comparing the fold increase in replication with Xrn1 knockdown from panel A or C to replication with siControl (no knockdown) treatment from panel B or D, respectively. The dotted line indicates 1-fold or no increase in replication due to knockdown. Significance was tested using ratio-paired t tests of relevant samples.