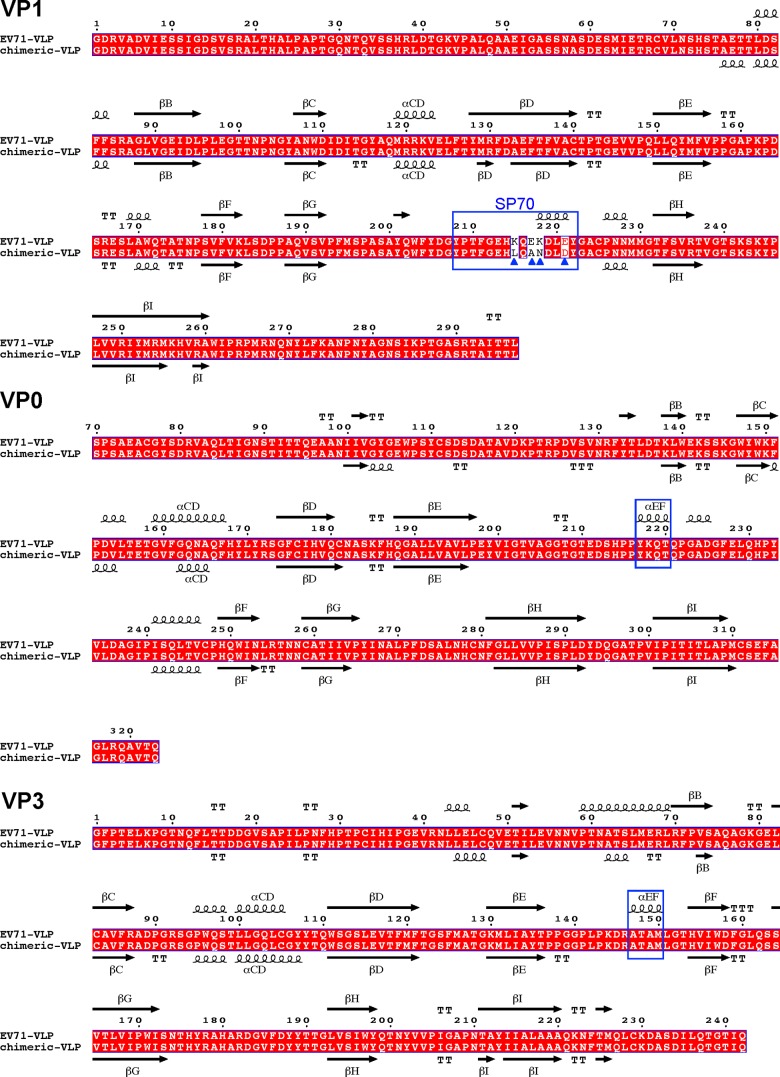
FIG 2.
Structure-based sequence alignments of EV71 VLP and EV71/CVA16 chimeric VLP. Capsid protein sequences used for the alignment include EV71 VLP and the chimeric VLP whose capsid proteins have been structurally determined. The secondary structure elements for EV71 VLP and chimeric VLP are shown at the top and bottom of the sequence alignment, respectively. The residue numbers correspond to those in EV71 VLP. Conserved residues are shown in white with a red background. Helices and strands are labeled according to the standard picornavirus nomenclature and are represented by coils and arrows, respectively. The blue triangles indicate the residues that are variable between EV71 VLP and the chimeric VLP. VP1 amino acids 208 to 222 corresponding to the SP70 region in EV71 VLP are boxed with a blue rectangle. VP0 residues 217 to 220, boxed with a blue rectangle, switched from a helical structure (the EF helix) in EV71 VLP to a loop conformation in the chimeric VLP. VP3 residues 147 to 150, boxed with a blue rectangle, changed from a helical structure (the EF helix) in EV71 VLP into a loop conformation in the chimeric VLP. This figure was produced using ESPript (33).