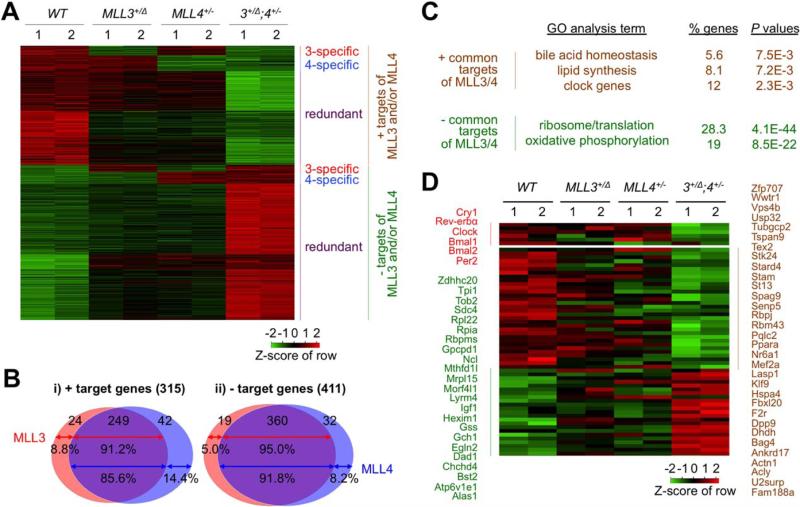
Fig. 2.
RNA-Seq assay identifies target genes of MLL3 and MLL4 in metabolism. (A) A heatmap shows that target genes of MLL3 and/or MLL4 uncovered by RNA-Seq analysis of liver mRNAs of 5-month-old WT, MLL3+/Δ, MLL4+/−, and MLL3+/Δ;MLL4+/− (n = 2) are grouped into six different clusters. (B) Venn diagram showing that a large portion of target genes of MLL3 and MLL4 (85.6%-95.0%) is commonly regulated by MLL3 and MLL4, whereas a much smaller portion of target genes of MLL3 and MLL4 (5.0%-14.4%) is regulated by MLL3 and MLL4 alone. (C) GO categories of the genes commonly regulated by MLL3 and MLL4. (D) A heatmap showing a subgroup of the target genes of MLL3 and MLL4, which have been previously shown to be regulated by MLL3 and to fluctuate in a circadian clock-dependent manner.16