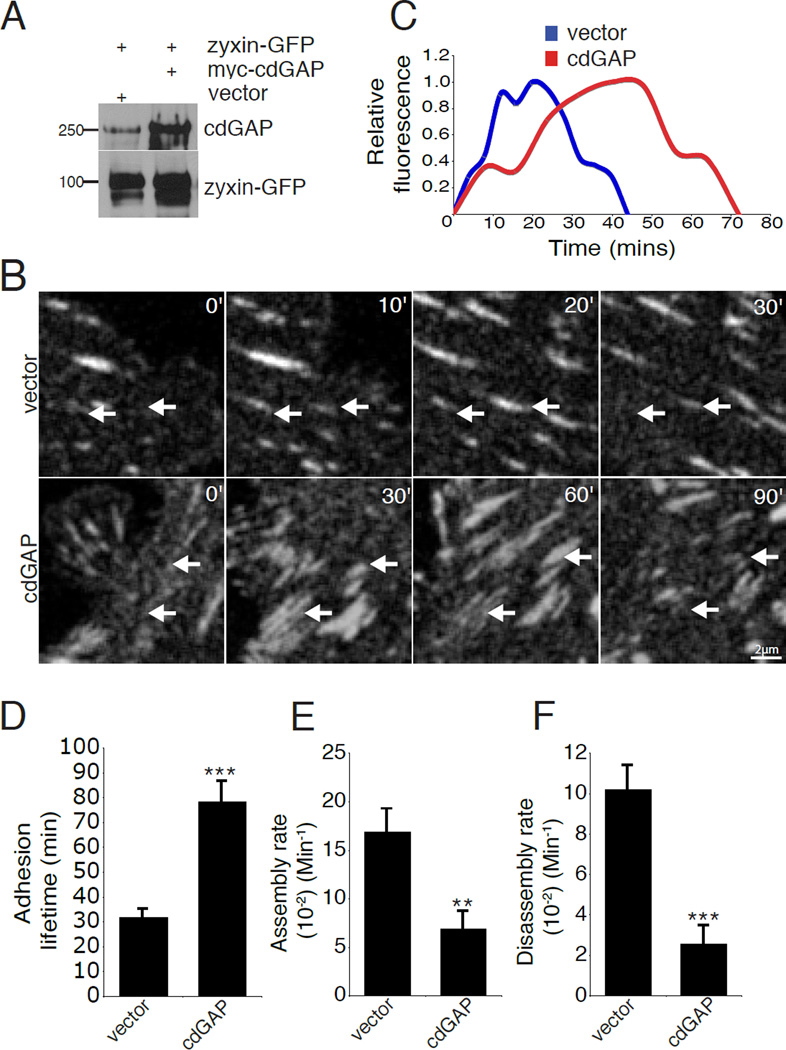
Figure 7. CdGAP over expression slows adhesion dynamics.
(A) U2OS cells were transiently co-transfected with zyxin-GFP and either vector or myc-cdGAP and evaluated for cdGAP and zyxin-GFP expression by western blot. (B) Zyxin-GFP positive U2OS cells expressing myc-control vector or myc-cdGAP were imaged to quantify focal adhesion dynamics. White arrows indicate adhesions that are turning over during the time course and labels indicate the duration of each sequence in minutes. (C) The relative fluorescence intensity of representative individual adhesions plotted over time. (D) Myc-cdGAP over expression lengthens adhesion lifetime. (E) Over expression of myc-cdGAP inhibits adhesion assembly rates. (F) Over expression of myccdGAP inhibits adhesion disassembly rates. Confocal image sequences were used to calculate the lifetime, adhesion assembly, and adhesion disassembly rates for each condition. All lifetime and rate measurements were made from four independent experiments for a minimum of 50 adhesions from 6–10 cells per condition. Statistical comparisons were made between control and myc-cdGAP over expressing cells for the lifetime and adhesion dynamics measurements with a Student’s t-test,**, p < 0.005 for adhesion assembly rate, *** , p < 0.001 for lifetime and disassembly rate comparisons. For Figure 7, all error bars represent +/− the standard error of the mean.