Fig. 4.
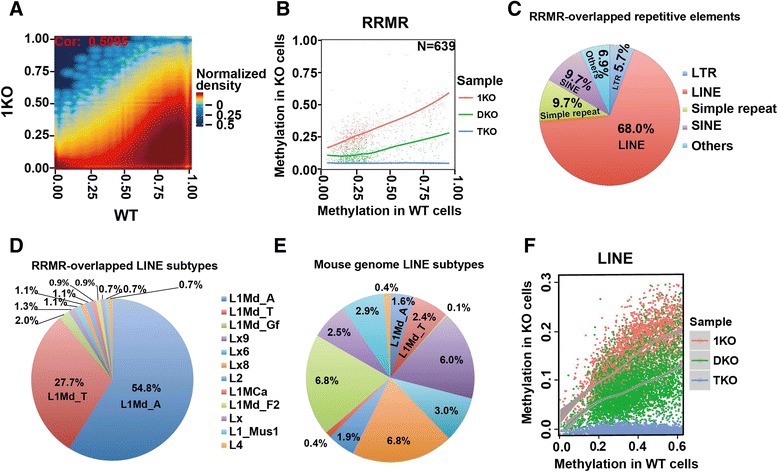
DNMT3a/3b-dependent (i.e., DNMT1 maintenance-independent) methylation patterns are critical for suppressing LINE retrotransposons. a Correlation of methylation in 1KO and WT cells. CG sites with coverage ≥10 in both cells were used. Color represents two-dimensional transformed density. b Methylation of RRMRs in three different ES KO cell lines relative to WT. c Overlap of 1KO cell RRMRs with repetitive elements in the mouse genome. SINE short interspersed nuclear element. d Overlap of 1KO cell RRMRs with LINE subtypes. Shown are percentages of each LINE subtype out of all LINE subtypes overlapping with RRMRs. e Percentages of different LINE subtypes in the mouse genome. f Methylation of LINEs in three different ES KO cell lines relative to WT
