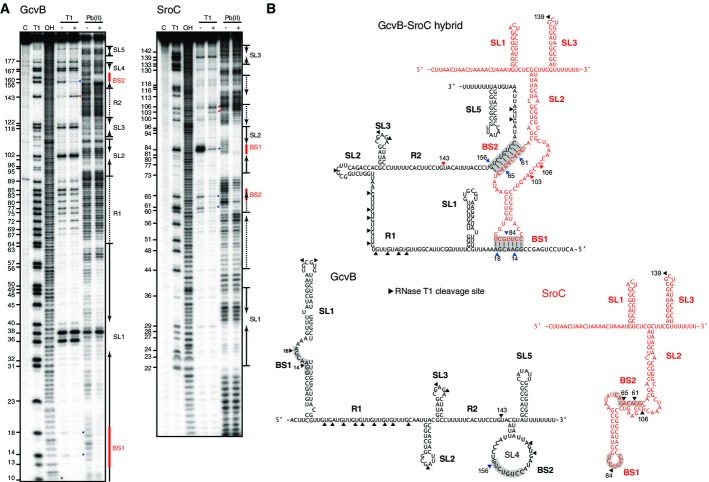
Figure 5.
- 20 nM of 5′-end-labeled GcvB and SroC were subjected to RNase T1 and lead(II) cleavage in the absence and presence of 100 nM cold sRNAs, SroC, and GcvB, respectively. Lane C: untreated RNA; lane T1: RNase T1 ladder of denatured RNA; lane OH: alkaline ladder. The position of G residues cleaved by RNase T1 is indicated at the left of picture.
- Secondary structures of GcvB and SroC, paired or alone. RNase T1 cleavage sites are indicated by arrowheads; color indicates those that appeared (red) or disappeared (blue) upon base-pairing. The base pairing sites BS1 and BS2 are shadowed.