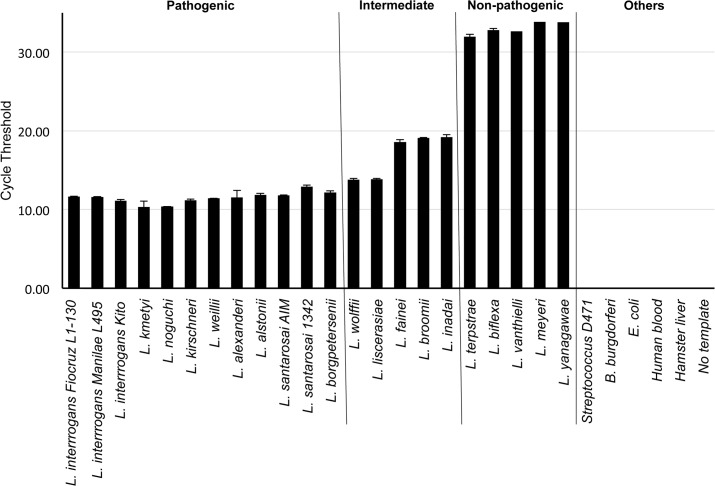
Fig 3. High sensitivity and specificity of 16S rRNA qPCR assay for pathogenic and nonpathogenic leptospiral species and serovars.
Total RNA was extracted from 17 high or intermediate pathogenic, and five non-pathogenic leptospiral species, Borrelia burgdorferi, group A Streptococcus, and Escherichia coli, as well as from uninfected human blood or hamster liver, as described in the materials and methods and converted into cDNA. Equal amount (10 ng) of cDNA templates from each bacterial species were subjected to qRT-PCR assays using 16S-1 primers and amplification cycles (Ct values) were measured. Note that the sensitivity of detection is the best for all tested highly pathogenic species or serovars followed by intermediate species while non-pathogenic strains display the lowest sensitivity (an average of 15 Ct values or 10,000 fold less detectability). All tested non-target bacterial species or mammalian samples remained undetectable. Data represent results from three independent experiments.