Summary
Gustatory receptors and peripheral taste cells have been identified in flies and mammals, revealing that sensory cells are tuned to taste modality across species. How taste modalities are processed in higher brain centers to guide feeding decisions is unresolved. Here, we developed a large-scale calcium imaging approach coupled with cell labeling to examine how different taste modalities are processed in the fly brain. These studies reveal that sweet, bitter, and water sensory cells activate different cell populations throughout the subesophageal zone, with most cells responding to a single taste modality. Pathways for sweet and bitter tastes are segregated from sensory input to motor output and this segregation is maintained in higher brain areas, including regions implicated in learning and neuromodulation. Our work reveals independent processing of appetitive and aversive tastes, suggesting that flies and mammals use a similar coding strategy to ensure innate responses to salient compounds.
Introduction
The sense of taste is primarily associated with feeding, allowing animals to identify food that is caloric and avoid toxic substances. Although feeding decisions are crucial for survival, little is known about the neural processing underlying taste acceptance or rejection in any organism. The taste system of Drosophila melanogaster affords an attractive model to study gustatory processing because it detects similar compounds and mediates similar behaviors as the mammalian gustatory system, but processing is carried out by an anatomically simpler nervous system that can be studied and manipulated with single cell precision.
Drosophila, like mammals, detects a few taste modalities that are innately tethered to food acceptance or rejection behavior (Liman et al., 2014; Yarmolinsky et al., 2009). Taste cells are found on the proboscis, legs, and wings of the fly (Stocker, 1994). The different classes of taste cells include bitter cells marked by the gustatory receptor GR66a and sweet cells marked by GR64f (Dahanukar et al., 2001; Dahanukar et al., 2007; Jiao et al., 2008; Marella et al., 2006; Thorne et al., 2004; Wang et al., 2004). In addition, water taste cells respond to low osmolarity, are inhibited by high osmolarity, and express the PPK28 ion channel (Cameron et al., 2010; Chen et al., 2010). These different classes of taste cells send axons to the subesophageal zone (SEZ) of the brain (Ito et al., 2014; Stocker, 1994). Motor neurons that drive feeding subprograms including proboscis extension and ingestion are also located in the SEZ (Gordon and Scott, 2009; Hampel et al., 2011; Manzo et al., 2012; Rajashekhar and Singh, 1994), suggesting that the SEZ may contain local circuits that process taste cues from detection to behavior. How different taste modalities are processed in the brain to guide feeding decisions is unresolved.
Studies of the mammalian gustatory system have led to two main models for how different taste modalities are encoded in the brain (Carleton et al., 2010; Lemon and Katz, 2007; Simon et al., 2006; Smith et al., 2000). The labeled line model of taste coding proposes that different taste modalities activate different pathways in the brain, leading to different behaviors. The mixed lines or across-fiber patterning model posits that information from modality-specific taste cells in the periphery becomes integrated in higher brain centers, such that single neurons respond to multiple modalities. In this model, the ensemble activity of broadly-tuned neurons or the dynamics of activation encodes taste quality. Recent studies in the mouse gustatory cortex and in the mouse geniculate ganglion, a primary relay from the tongue, support a labeled line model (Accolla et al., 2007; Barretto et al., 2015; Chen et al., 2011).
Here, we examined how mammalian models of taste coding apply to the simpler gustatory system of the fly, to gain insight into successful coding schemes used through evolution. To examine gustatory processing, we developed a large-scale calcium imaging approach to monitor neural responses throughout the SEZ with cellular resolution. This allowed us to examine the neurons activated by different taste modalities and taste mixtures. Our studies show that bitter and sweet modalities are processed by different cells and demonstrate segregated processing for appetitive and aversive tastes.
Results
Reproducibility of taste responses in the SEZ
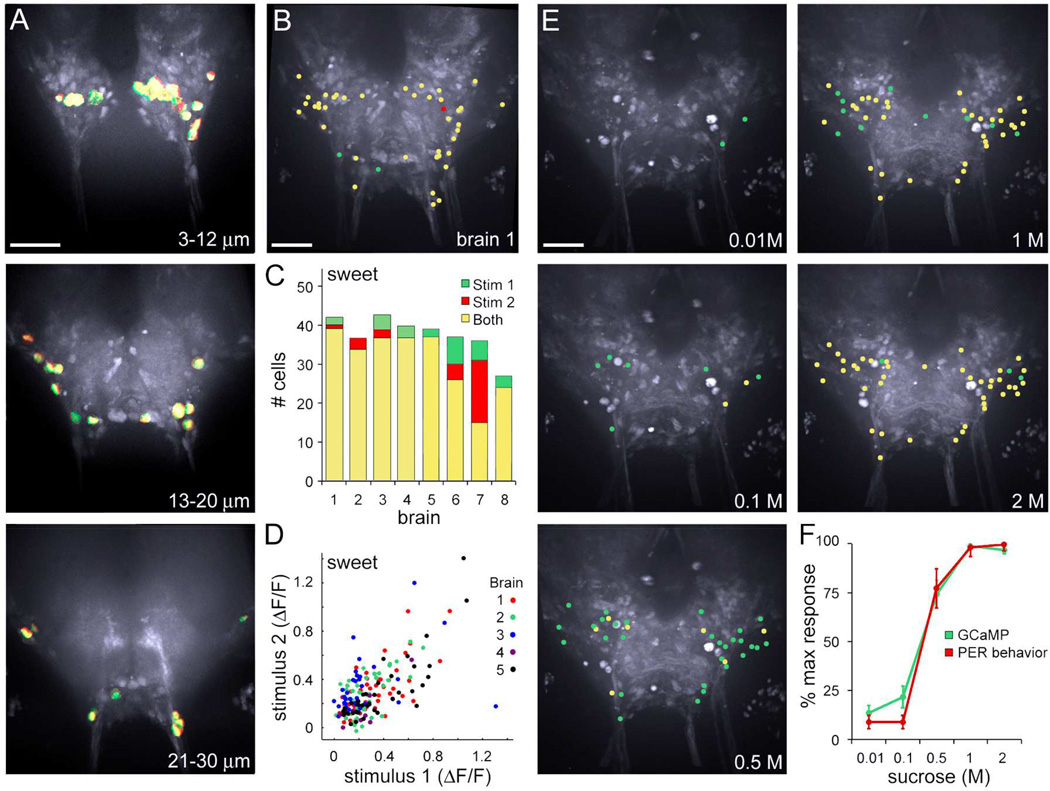
To monitor activity throughout the SEZ, we expressed the genetically encoded calcium indicator GCaMP6s, which has high sensitivity, low basal fluorescence, and slow decay (Chen et al., 2013), using the pan-neural nSynaptobrevin-Gal4 promoter (Pauli et al., 2008). Proboscis taste cells of a live fly were stimulated and fluorescence changes in the brain were monitored with spinning disk confocal microscopy to serially monitor the SEZ depth, by continuously scanning 23 Z-sections, 87ms/1.3µm section (Figure 1A, see Movie S1–S4 for raw and processed movies, Figure S1 for data analyses). A nuclear marker (Pandey et al., 2005) was included to determine fluorescence changes in single cells (Figure 1B–C). The data from all z-sections were used in the analysis in order to identify responsive neurons (see Methods, Image Acquisition). For visualization purposes, cellular fluorescence changes of responding cells were then compressed into a single image (Figure 1D), a few thick sections (other Figures), or displayed as schematic representations for the whole SEZ (Figure 1E). This approach allowed rapid monitoring of neural activity throughout the SEZ.
Figure 1. Monitoring activity throughout the entire fly SEZ.
A. (left) Schematic of the fly brain showing the SEZ and imaging area (boxed). To monitor activity, 23 1.3µm Z-sections were scanned at 0.5 Hz/Z-stack throughout the entire SEZ of a living fly pre- and post- taste delivery to the proboscis (right). Image of the fly brain was modified from http://www.flybrain.org.
B. One Z-section with automated ROIs (outlined in white) marking cell nuclei based on histone-RFP (red) with maximum ΔF response (green) to the 2M sucrose taste stimulus overlaid. Scale is 50 µm.
C. Z-section in B, showing cell bodies (green, outlined in white) with maximum ΔF significantly above background (2 st dev) and the nuclear marker, histone-mRFP (red).
D. Maximum ΔF of flattened Z-sections representing the entire SEZ, showing activated cell bodies overlaid on average maxF image.
E. Schematic representing all responding cells in SEZ (green circles) overlaid on average maxF image. See Movies S1–S4 for raw and processed GCaMP responses and Figure S1 for ΔF responses of single cells and different frame rates.
We used a combination of exogenous activation and natural compounds to specifically activate different classes of gustatory sensory neurons. High osmolarities were used to prevent water cell activation (Cameron et al., 2010) and allow independent activation of sweet, bitter, and water sensory cells. The ATP-gated cation channel P2X2 (Lima and Miesenbock, 2005) was expressed in sweet-sensing neurons (Gr64f-LexA), bitter-sensing neurons (Gr66a-LexA), or water-sensing neurons (ppk28-LexA) (Miyamoto et al., 2013; Thistle et al., 2012), and the proboscis was stimulated with 100mM ATP for specific activation of these sensory classes. P2X2-based activation provided a means to strongly and reproducibly activate specific classes of sensory cells, with little or no desensitization across trials. In addition, the proboscis was stimulated with 2M sucrose to selectively activate sweet sensory neurons and enable cross-modality comparisons. These stimuli triggered responses in central neurons throughout the SEZ.
Overlaying the maps of responding cells for two sequential stimulations (trial 1 in green, trial 2 in red) illustrates the reproducibility (yellow), as seen in fluorescence images (Figure 2A, Figure 3A, Figure S2A,), schematic representations (Figure 2B, Figure 3B, Figure S2B), and cell counts (Figure 2C, Figure 3C, Figure S2C). In addition, plots comparing the maximum fluorescence change of each cell for trial 1 versus trial 2 illustrates that response magnitudes are quantitatively similar across trials (Figure 2D, Figure 3D, Figure S2D).ATP-mediated stimulation of sensory cells and high tastant concentrations activated the same populations (Figure 2C, brains 6–8; Figure 3C, brains 6–8), arguing that exogenous activation evoked saturating responses. Consistent with this, the number of sucrose-responsive cells was concentration-dependent and plateaued at high concentrations, mirroring behavioral responses to sucrose (Figure 2EF). The number of bitter-responsive cells showed a similar concentration-dependence (Figure 3EF), although activation of bitter sensory neurons expressing P2X2 provided the strongest and most reliable responses. The repeatability was 88 ± 1% across all two tastant stimulation trials, representing 15 animals (Figure 2, Figure 3, Figure S2), with 34 ± 1 bitter-responsive, 39 ± 1 sucrose-responsive and 15 ± 1 water-responsive cells/SEZ. These experiments demonstrate that taste cell stimulation generated reliable and reproducible activation of central cell populations.
Figure 2. Sucrose stimulation elicits reproducible and concentration-dependent cell activation in the SEZ.
A. Cells responding to 2M sucrose stimulation of the proboscis, with responses to the first (green) and second (red) stimulation overlaid. Images are maximum ΔF of cell bodies in flattened Z-sections (representing anterior 3–12 µm, middle 13–20 µm, and posterior 21–30 µm). Scale is 50 µm.
B. Schematic showing all active cells in SEZ, with yellow representing cells activated by two sequential stimulations and red/green by one stimulation for brain in A (brain 1 in C).
C. Summary of responding cells for eight brains. Brains 1–5 represent sequential stimulations of 2M sucrose. Brains 6–8 represent one stimulation of 2M sucrose (green) and one stimulation of sweet sensory cells using ATP-induced activation of GR64f cells expressing P2X2 (red) and cells responding to both (yellow).
D. Plot of single cell responses (ΔF/F) for two sequential sucrose stimulations for brains 1–5 in C, showing magnitudes of fluorescence changes across trials.
E. Stimulation of the fly proboscis with increasing sucrose concentrations (0.01, 0.1, 0.5, 1, 2 M) progressively recruited more taste-responsive cells in the SEZ, shown as schematics of responding cells at the different concentrations. Green denotes cells that responded at the given concentration but not lower concentrations, yellow denotes cells that also responded at a lower concentration.
F. The fraction of responding cells by GCaMP calcium imaging and the probability of proboscis extension behavior (PER) showed the same sucrose concentration dependence. Responses are normalized to maximum response. GCaMP (green) is mean ± SEM for 5 brains. Proboscis extension response (red) is 3 trials, n=10 flies/trial, mean ± SEM. Responsiveness in the imaging preparation is reduced as compared to intact flies. See Figure S2 for reproducible activation of central cells to water sensory cell activation.
Figure 3. Reproducibility and dose dependence of bitter responses.
A. Cells responding to ATP-induced activation of proboscis GR66a bitter cells expressing the ATP-gated channel P2X2. Responses to the first (green) and second (red) stimulation are overlaid. Images are maximum ΔF of cell bodies (anterior 3–12 µm, middle 13–20 µm, and posterior 21–30 µm). Scale is 50 µm.
B. Schematic showing all active cells in SEZ, with yellow representing cells activated by two sequential stimulations and red/green by one stimulation for brain shown in A (brain 1 in C).
C. Summary of responding cells for eight brains. Brains 1–5 represent sequential stimulations with a mixture of bitter compounds and ATP. Brains 6–8 represent one stimulation with bitter compounds (green) and one stimulation with ATP for P2X2-mediated activation of GR66a cells (red) and cells responding to both (yellow).
D. Plot of single cell responses (ΔF/F) for two sequential bitter stimulations for brains 1–5 in C, showing magnitudes of fluorescence changes across trials.
E. Stimulation of the fly proboscis with increasing denatonium concentrations (0.5, 1, 10, 100 mM). Red denotes cells that responded at the given concentration but not lower concentrations, yellow denotes cells that also responded at a lower concentration. Activation of proboscis GR66a bitter cells expressing the ATP-gated channel P2X2 with ATP and a bitter mix activated the most neurons reliably but many of these are also activated by bitter stimuli.
F. The number of responding cells by GCaMP calcium imaging increased with concentration. Mean ± SEM for 5 brains.
Taste quality representations in the SEZ
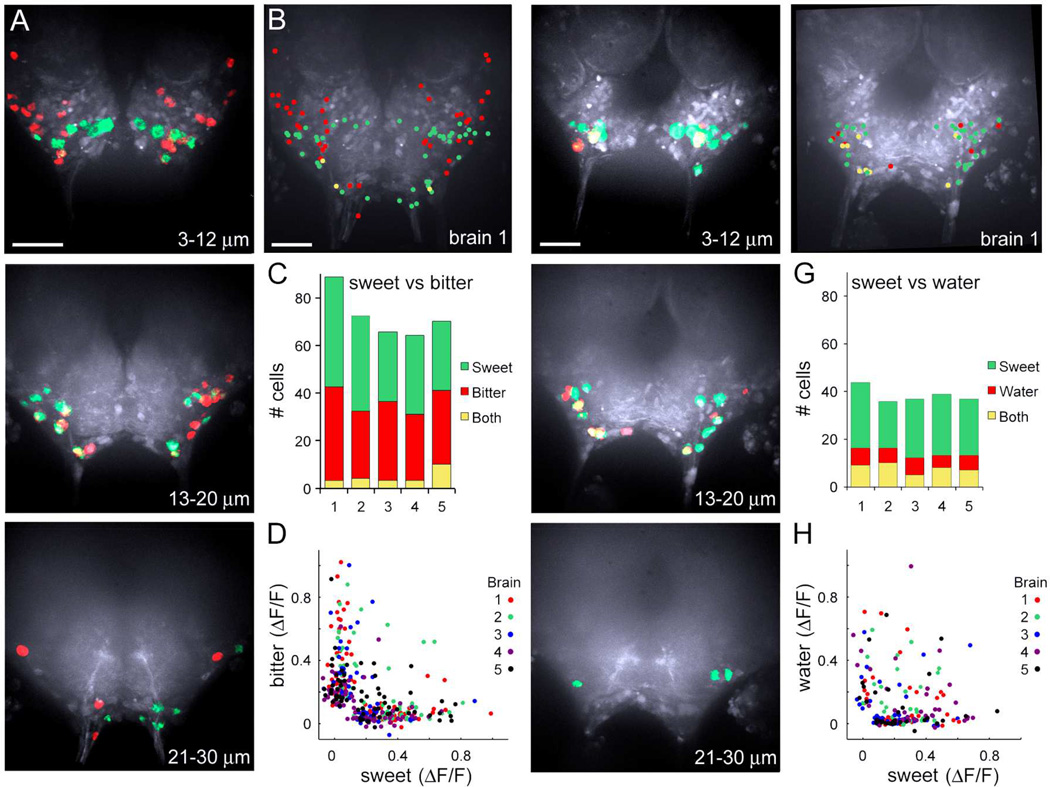
This large-scale calcium imaging approach enabled us for the first time to ask whether Drosophila higher-order gustatory neurons are tuned to a single taste modality or are broadly tuned across modalities. Different gustatory sensory classes were sequentially activated and the responses were mapped onto the SEZ for within-brain comparisons. Comparing the response to 2M sucrose and ATP-mediated activation of bitter cells revealed that these taste modalities activated different cells (Figure 4A–D), with 36 ± 4 sucrose-selective, 32 ± 2 bitter-selective, and 5 ± 1 cells/SEZ activated by both compounds. Similarly, water and bitter sensory stimulation activated different cells, with only 4 ± 1 of the 34 ± 3 cells responding to both compounds (Figure S3A–D). This demonstrates that the vast majority of higher-order taste cells do not respond to all taste modalities and shows that bitter sensory stimulation activates different cells than sucrose or water stimulation.
Figure 4. Taste quality maps in the SEZ.
A. Cells responding to ATP-induced activation of proboscis bitter cells expressing P2X2 (red) and cells responding to 2M sucrose (green). Images are maximum ΔF (representing anterior 3–12 µm, middle 13–20 µm, and posterior 21–30 µm). Scale is 50 µm.
B. Schematic showing all active cells in SEZ, with red representing cells activated by bitter taste cell activation and green by sucrose stimulation for brain shown in A. Yellow represents cells responding to bitter and sucrose stimulation (brain 1 in C).
C. Summary of responding cells for five brains. Cells responding to ATP-induced activation of GR66a bitter cells expressing P2X2 (red), cells responding to 2M sucrose (green) and cells responding to both (yellow).
D. Plot of single cell responses (ΔF/F) to bitter stimulations versus sucrose stimulations for brains 1–5 in C, showing modality-selective responses.
E. Cells responding to ATP-induced activation of proboscis PPK28 water-sensing cells expressing P2X2 in red and cells responding to 2M sucrose in green.
F. Schematic showing all active cells in SEZ, with red representing cells activated by water taste cell activation and green by sucrose stimulation for brain shown in E. Yellow represents cells responding to water and sucrose stimulation (brain 1 in G).
G. Summary of responding cells for five brains. Cells responding to ATP-induced activation of PPK28 water-sensing cells expressing P2X2 (red), cells responding to 2M sucrose (green) and cells responding to both (yellow).
H. Plot of single cell responses (ΔF/F) to water stimulations versus sucrose stimulations for brains in G. See Figure S3 for a comparison of water versus bitter-responsive cells.
To further test whether higher-order neurons are tuned to single taste modalities, we compared the responses to sucrose and water cell activation, two appetitive taste stimuli that trigger feeding initiation in the fly. Again, we observed that many central neurons responded selectively to a single taste quality and some cells responded to both compounds. On average, 23 ± 1 cells/SEZ were sucrose-selective, 6 ± 1 were water-selective, and 8 ± 1 cells were activated by both 2M sucrose and water cell stimulation (Figure 4E–H).. More cells were activated by both sucrose and water than by other taste pairs (Figure S3EF, t-test, **P<0.01), suggesting that these cells might represent the convergence of sweet and water modalities onto a pathway for taste acceptance behavior. Together, these studies show that most cells in the SEZ respond to single taste modalities and a few cells respond to multiple modalities.
Taste mixtures do not activate additional cells
Although the responses to single taste compounds suggest that central neurons are not broadly tuned across all taste modalities, one caveat is that multimodal cells might only weakly respond to single compounds but may respond more strongly to combinations. If this were the case, then we would expect that taste mixtures should activate more cells than single compounds. To test this, flies were stimulated with 2M sucrose alone, then bitter cell activation alone to generate a map of sweet- and bitter-responsive cells (Figure 5A). Flies were then stimulated with a mixture of the sweet and bitter solutions (same final concentration), and the responding cells were color-coded based on the response to single compounds (Figure 5B). Few additional cells were activated by the mixture (0.3 ± 0.3 cells per brain) that were not activated by single compounds alone, consistent with trial-to-trial variability and arguing against large populations of broadly-tuned cells.
Figure 5. Mixtures do not activate additional cells.
A. Schematic showing taste-responsive cells in SEZ, with cells responding to bitter cell activation in red, responding to 2M sucrose stimulation overlaid in green and cells responding to both (yellow) (Separate 1 in C). Scale is 50 µm.
B. Schematic showing cells responding to the mixture of bitter cell activation + 2M sucrose, with responding cells color-coded based on the response to single compounds. Cells that responded to the mix only are blue (Mix 1 in C).
C. Number of cells responding to individual compounds (Separate) versus the mixture (Mix) for three brains (1,2,3). Color-coding for mixtures is based on the response to single compounds.
D. Average number of cells responding to individual compounds (Separate) versus the mixture (Mix), color-coded based on response to single compounds for the 3 brains in C, mean ± SEM, paired t-test, ***p<0.001, **p<0.01.
E–H. Comparison of responses to single compounds versus mixtures for 2M sucrose and ATP activation of water sensory cells expressing P2X2, showing a schematic sample brain for single compounds (E), mixture (F), summary graph for 3 brains (G), and average number of cells responding for 3 brains in G, mean ± SEM, paired t-test, *p<0.05 (H). Cells responding to ATP-induced activation of PPK28 water-sensing cells (red), cells responding to 2M sucrose (green), cells responding to both (yellow) and cells responding to mix (blue).
I. ΔF/F image of sensory axons responding to 500mM sucrose.
J. ΔF/F image of sensory axons responding to 500mM sucrose plus 10mM denatonium. Same brain as I.
K. Suppression is concentration-dependent. (left) Fluorescence changes in sensory axons decreased with increasing denatonium concentration. ΔF/F response is normalized to 500mM sucrose response. n=5; mean ± SEM. (middle) Number of sucrose-responsive cells in SEZ decreased as denatonium increased, while number of non-sucrose-responsive cells increased (bitter-responsive cells). n=5; mean ± SEM. (right) Proboscis extension to 500mM sucrose decreased with inclusion of denatonium. 3 trials, n=10 flies/trial, mean ± SEM. See Figure S4 for suppression of sucrose responses in the presence of a GABAB receptor agonist.
In fact, instead of activating additional cells, mixtures activated far fewer cells than the single components, revealing mixture suppression (81 ± 5% of sweet-responsive cells and 34 ± 9% of bitter-responsive cells were suppressed) (Figure 5CD). Mixture suppression also occured for water and sucrose, two appetitive stimuli, based on sequential and simultaneous activation of these two sensory classes (94 ± 6% of water-responsive cells and 36 ± 9% of sucrose-responsive cells were suppressed) (Figure 5E–H). Plots of single-cell fluorescence changes to mixtures relative to single compounds revealed general decreased responses (Figure S4AB). Thus, taste mixtures suppressed central responses, suggesting cross-inhibition between different sensory pathways.
Recent studies of gustatory sensory responses revealed that bitter compounds inhibit the activity of sweet sensory cells (Chen and Amrein, 2014; Chu et al., 2014; Jeong et al., 2013), suggesting that cross-inhibition at the level of the sensory neuron might cause the reduction we observed in the central response to taste mixtures. Indeed, sensory axons that responded to 500 mM sucrose stimulation showed a dose-dependent activity decrease in the presence of the bitter compound denatonium, consistent with recent studies (Figure 5I–K) (Chu et al., 2014). Similarly, the activity of sweet-sensing neurons was suppressed by water cell activation (12 ± 3% reduction, n=6, t-test to two sequential sucrose activations, P<0.01). We also found that suppression of sucrose-responsive cells in the SEZ showed a similar dose sensitive suppression by bitter sensory activation, which mirrored bitter suppression of the behavioral response to sucrose (Figure 5K). The GABAB receptor expressed in sweet sensory neurons mediates presynaptic inhibition in the presence of bitter compounds, with the GABABR agonist SKF97541 acting to reduce sensory activity (Chu et al., 2014). Notably, we found that SKF97541 also strongly suppressed the activity of central sucrose-responsive cells (Figure S4C–F). These studies are consistent with the notion that reduced sensory activity contributes to mixture suppression. Furthermore, they show that mixtures adjust the relative activation of the sweet versus bitter pathways rather than activate additional multimodal cells.
Taste stimulation activates proboscis motor neurons
To test whether the cells responding to taste stimulation indeed represent key elements of gustatory circuits, we examined whether the responding cells include motor neurons (MNs) known to drive feeding programs. We generated flies that co-expressed a photoactivatable GFP (Ruta et al., 2010) along with GCaMP6s in all neurons in order to monitor taste-induced activity and then label proboscis MNs (Figure 6). Photoactivation of C3PA-GFP in the nerves carrying MN axons reliably backfilled the majority of MNs previously identified by anatomical studies (30 MNs by anatomical studies; 19±1 MNs by pa-GFP) (Rajashekhar and Singh, 1994). Labeling by pa-GFP identified 12±1 sucrose-responsive MNs and 24±1 non-labeled sucrose-responsive cells (Figure 6D and Figures S5A, showing single Z-section overlay), demonstrating that calcium imaging reports activation of circuits from sensory input to motor output.
Figure 6. Sensory activation elicits motor responses.
A. Maximum ΔF of cells responding to sucrose stimulation, with cell bodies overlaid on average maxF image. Scale is 50 µm. (top) Motor neurons filled by photoactivation of C3PA-GFP in proboscis nerves (labelled red). (middle) Overlay showing the sucrose-responsive neurons that are motor neurons (MNs). (bottom). See Figure S5A for a single Z-section.
B. Schematic showing all active cells in SEZ, with sucrose-responsive cells not labelled by C3PA-GFP (green), sucrose-responsive motor neurons (yellow), and non-responsive filled motor neurons (red) for brain in A (brain 1 in D).
C. Schematic for brain 2 in D.
D. Summary of sucrose-responsive cells not labelled by C3PA-GFP (green), sucrose-responsive motor neurons (yellow), and non-responsive filled motor neurons (red) for five brains.
E. SEZ taste responses were monitored in DVGLUT-Gal4, UAS-GCaMP6s flies, which express GCaMP6s in glutamatergic motor neurons and interneurons. Brain (brain 1 in G) showing VGLUT cells activated by sucrose stimulation (green) or bitter sensory cell activation (red).
F. Second sample brain (brain 2 in G) showing VGLUT cells activated by sucrose stimulation (green) or bitter sensory cell activation (red). The apparent overlap stems from compressing a 3D representation into 2D (the red cells are below the green cells and not-overlapping).
G. Summary of VGLUT taste-responsive cells from 5 brains, showing segregation of bitter and sucrose candidate MNs. See Figure S5 for pan-neural calcium imaging with VGLUT cell nuclei labeled, showing activation of VGLUT and non-VGLUT cells.
As an alternative approach to identify taste-responsive MNs, we monitored SEZ taste responses in DVGLUT-Gal4, UAS-GCaMP6s flies, which express GCaMP6s in glutamatergic motor neurons and interneurons (Daniels et al., 2008). Sucrose stimulation activated 13±2 DVGLUT cells, whereas bitter cell stimulation activated a non-overlapping set of 6±1 DVGLUT cells (Figure 6E–G). In addition, monitoring pan-neural calcium activity with nSyb-LexA, lexAop-GCaMP6s flies expressing nuclear histone-RFP in DVGLUT cells identified 12±1 sucrose-responsive DVGLUT cells and 25±1 non-labeled sucrose-responsive cells, consistent with pa-GFP studies (Figure S5B–E). These studies argue that sweet and bitter activate different motor neurons, consistent with behavioral studies that show that flies extend the proboscis to sugars and retract it to bitter compounds (Dethier, 1976). In addition, these studies show that the majority of taste-responsive cells in the SEZ are not motor neurons but rather modality-selective interneurons.
Two specific MN classes in the SEZ have previously been shown to respond to sugars but not bitter compounds: E49 MN that drives proboscis extension and MN11+12 that drive ingestion (Gordon and Scott, 2009; Manzo et al., 2012). We tested whether these cells are consistently detected by pan-neural imaging, by monitoring calcium activity with nSyb-LexA, lexAop-GCaMP6s while expressing tdTomato in the each cell class. Both classes showed robust responses to sucrose stimulation (Figure S6), demonstrating that pan-neural imaging reliably detects known taste-responsive MNs and arguing for the underlying stereotypy of taste circuits.
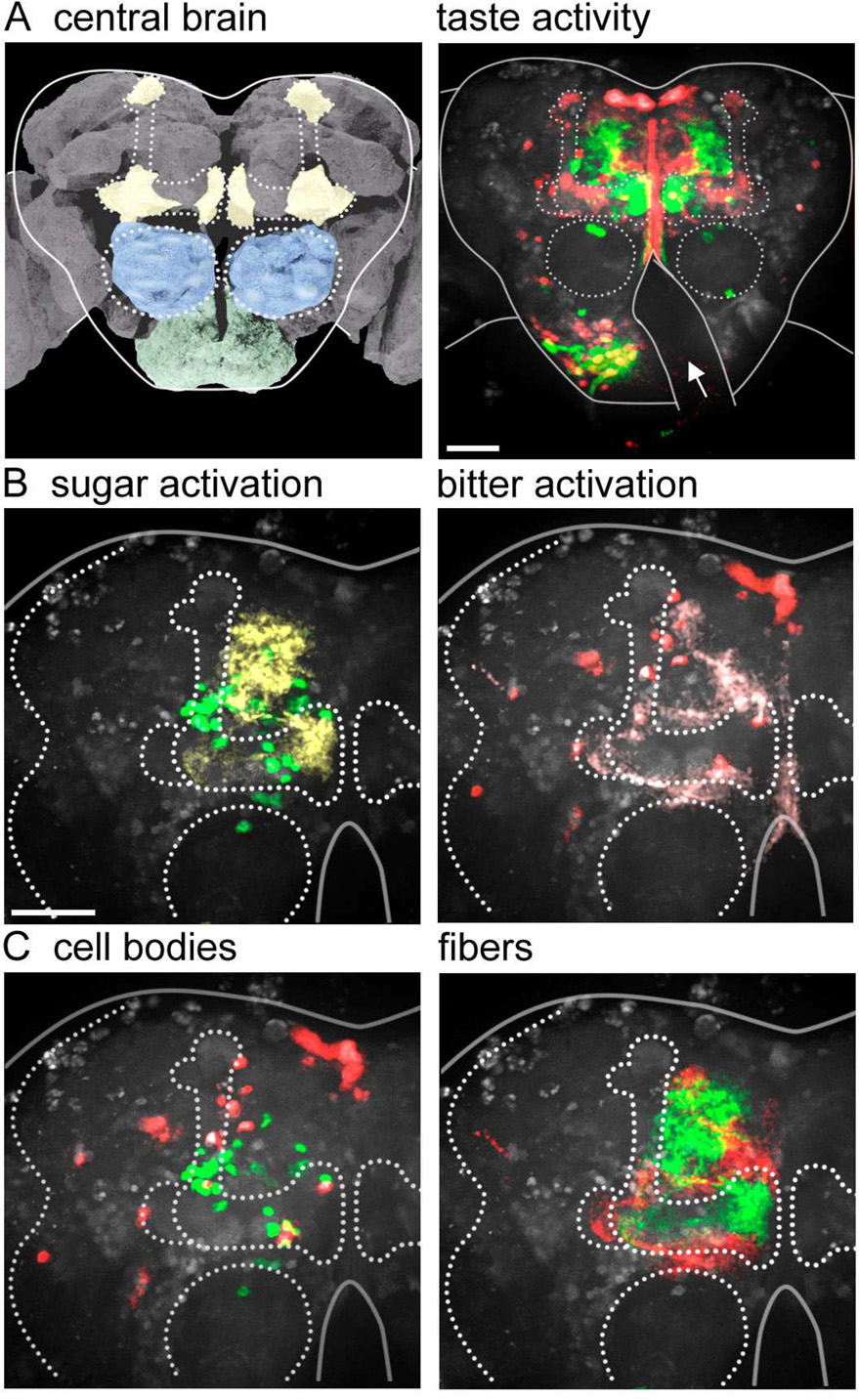
Taste representations in higher brain centers
To examine whether appetitive and aversive tastes are processed in separate pathways in higher brain regions as well as in the SEZ, we stimulated different classes of proboscis taste neurons and monitored activity in the anterior shell of the higher brain. Stimulation of sweet or bitter sensory cells produced activity in sparse and distributed cell populations in these higher centers (Figure 7). In both cases, prominent activation occured in the lobes of the mushroom bodies (MBs), the site for associative learning in the fly (Heisenberg, 2003). Previous studies have shown that a subset of MB extrinsic neurons, PAM neurons, respond to sucrose sensory stimulation and convey reward (Burke et al., 2012; Liu et al., 2012). Pan-neural imaging in the background of labeled PAM neurons confirmed that PAM activity is consistently detected in the higher brain upon sucrose stimulation (Figure S6), providing further validation of the pan-neural imaging approach. The observation that sweet and bitter stimulation activated MBs is consistent with their role as unconditioned stimuli in associative learning paradigms. Our calcium imaging demonstrates that taste-induced activity is compartmentalized in the MB region, with bitter and sweet activating non-overlapping regions.
Figure 7. Sweet and bitter activate different pathways in higher brain regions.
A. (left) Schematic of fly brain, showing mushroom bodies (yellow), antennal lobes (blue) and SEZ (green). Image modified from http://www.flybrain.org. (right) Taste-induced activity in the anterior shell of the fly brain, with responses to sucrose sensory stimulation in green and bitter in red. Dotted lines highlight mushroom bodies and antennal lobes (circles). Solid lines outline the central brain, with arrow noting obstruction by the esophagus. Pars intercerebralis is the top group of cells at the midline. Scale is 50 µm.
B. Higher resolution view of the upper left quadrant of the fly brain, different brain than A, with sucrose-responsive cell bodies in green and projections in yellow. On right are bitter-responsive cells in red and projections in pink. Scale is 50 µm.
C. Comparison of sucrose-responsive (green) and bitter-responsive (red) cell bodies (left) or projections (right). Images are representative of n=5 brains.
In addition to strong activation in the MBs, taste compounds activated scattered cells in the protocerebrum. Bitter compounds also activated pars intercerebralis neurons, a major neurosecretory center in the brain (Nassel, 2002). The activation pattern in the higher brain suggests that taste circuits are relatively sparse. Importantly, in these higher brain centers, sweet and bitter stimulation still activated different cells, demonstrating separation of taste processing pathways in the higher brain.
Discussion
A central question in taste processing is how different taste modalities are encoded in the brain. In the mammalian gustatory system, labeled lines, mixed lines, and temporal dynamics have all been proposed as fundamental stategies used by the nervous system to process tastes, with recent evidence favoring labeled line encoding (Accolla et al., 2007; Barretto et al., 2015, Chen et al., 2011, Smith et al., 2000). In Drosophila, the limited understanding of neural circuits beyond sensory and motor neurons has precluded examination of modality processing. Here, we take advantage of recent improvements in genetically-encoded calcium indicators and high-speed, multi-plane imaging to examine taste-induced activity throughout the SEZ and anterior brain, similar to whole-brain calcium imaging approaches in zebrafish and C. elegans (Ahrens et al., 2012; Ahrens et al., 2013; Schrodel et al.). This approach enabled us to examine the neurons activated by different taste modalities and probe models of taste coding.
Monitoring brain activity in response to stimulation of different gustatory classes revealed that the majority of central neurons responded selectively to bitter, sweet, or water sensory cell activation. In addition, some neurons responded to both water and sucrose and may represent positive valence or taste acceptance behavior, while a few responded to other taste pairs. This demonstrates that most central taste-processing neurons in the fly do not respond to multiple taste modalities and argues against models of taste coding based on broadly-tuned cells. Instead, we find that taste representations are largely modality-specific, indicating separate processing streams for different taste qualities. Moreover, bitter and sweet stimulation activate different sets of proboscis motor neurons and activate different neurons in the higher brain. This argues that sweet and bitter tastes are processed by segregated pathways, suggesting a strategy that ensures innate responses to essential compounds.
Examining responses to taste mixtures in the SEZ revealed that no additional cells are activated by mixtures that are not activated by single components, again arguing against nonselective cells. Instead, mixtures activated only a fraction of the cells that responded to the individual components. Mixture suppression occured for sugar/bitter mixes and sugar/water mixes, demonstrating inhibition between appetitive and aversive stimuli as well between two appetitive stimuli. Recent studies have identified two mechanisms by which bitter compoiunds inhibit sweet sensory activation: (1) bitter compounds bind a chemosensory binding protein which acts on sweet sensory cells to inhibit activity (Jeong et al., 2013) and (2) bitter compounds activate GABAergic neurons, causing GABAB receptor-mediated pre-synaptic inhibition of sweet sensory axons (Chu et al., 2014). Our study shows that taste mixtures reduce sensory activation and reduce the number of taste-responsive cells in the SEZ, consistent with the notion that decreased sensory activity contributes to mixture suppression. Cross-inhibition of different gustatory modalities is an effective strategy to compare the ratio of sweet to bitter and adjust the relative activation of acceptance versus avoidance pathways.
This study represents the first large-scale analysis of pan-neural activity in the fly brain. The advantages of this approach are that it is possible to monitor the activity of large populations at single-cell resolution which is not feasible by other approaches, it enables unbiased sampling that does not require specific Gal4 lines expressed in already-known cells of interest, and it provides rapid evaluation of brain-wide activity. Whereas previous studies had identified four classes of taste-responsive cells (Burke et al., 2012; Flood et al., 2013; Gordon and Scott, 2009; Liu et al., 2012; Manzo et al., 2012), our study uncovered more than 100 taste-responsive cells, the vast majority of which are modality-specific. As this approach relies on calcium imaging in the soma, the sensitivity of GCaMP6, and the expression levels driven by nSynaptobrevin-Gal4, detection limitations may exist. Nevertheless, this study provides a population overview of gustatory processing in the fly brain and a framework for future studies to determine the anatomy and connectivity of taste-responsive neurons.
We show here that different taste modalities in the periphery activate different pathways in the brain, consistent with labeled line taste processing. Information is processed in separate streams for appetitive and aversive tastes, which are maintained in the higher brain and are mutually inhibitory. Recent studies in the mammalian gustatory system argue for modality-specific representations in the gustatory cortex and are consistent with the labeled line model (Accolla et al., 2007; Barretto et al., 2015; Chen et al., 2011; Schoenfeld et al., 2004; Yamamoto et al., 1985), suggesting that dedicated pathways may be a general strategy to process tastes used throughout evolution.
Experimental Procedures
Experimental Animals
Drosophila stocks were maintained on standard cornmeal/agar/molasses medium at 22°C. The following transgenes were used: nSyb-Gal4 (Pauli et al., 2008); UAS-GCaMP6s (Chen et al., 2013); His2Av-mRFP (Pandey et al., 2005); LexAop-P2X2 (Yao et al., 2012); Gr66a-LexA::VP16, ppk28-LexA::VP16 (Thistle et al., 2012); Gr64f-LexA::VP16 (Miyamoto et al., 2013); UASC3PA-GFP (Ruta et al., 2010); VGLUT-Gal4 (Daniels et al., 2008); E49-Gal4 (Gordon and Scott, 2009); MN11+12-Gal4 (Manzo et al., 2012); R58E2-LexA (the PAM driver line) (Liu et al., 2012); lexAop-GCaMP6s (Douglas Kim, Janelia Research Campus); UAS-CD8-tdTomato (Thistle et al., 2012); lexAop-myrCherry (Diegelmann et al., 2008).
G-CaMP Imaging in the SEZ
Female flies, 2–3 days old, were food deprived on a wet kimwipe for 18–24 hrs. They were briefly anesthetized using CO2 and placed into a small slit on a custom-built plastic mount at the cervix such that the head was isolated from the rest of the body. The head was then immobilized using nail polish and the proboscis was waxed at the maxillary palps in an extended position to allow for taste solution delivery. A coverglass was placed at the base of the rostrum at a 45-degree angle to the plane of the plastic mount such that the proboscis was isolated from the head. The head cuticle was dissected in ice-cold adult hemolymph-like solution (AHL) (Wang et al., 2003) lacking calcium and magnesium, the air sacs were removed, and the esophagus was severed to allow viewing of the SEZ. Calcium- and magnesium-free AHL was replaced by AHL prior to imaging.
For imaging the anterior shell of the higher brain, the remaining head cuticle, air sacs and debris were removed using fine forceps. The eyes were removed to limit vision-dependent neural activity. The esophagus was kept intact, as that generated more reliable responses in higher brain areas.
Tastant Stimulations
High tastant concentrations were used to ensure maximal responses in order to identify as many taste-responsive cells as possible, essential for the goals of the study. These concentrations were validated in a number of ways:
Dose-response curves identified maximal activation and showed that many of the cells responding at the highest concentration responded at lower concentrations, arguing against transformation of the response at higher concentrations.
Sucrose and bitter concentrations used are consistent with calcium imaging studies from other laboratories studying taste activity (Chu et al, Current Biology 2014; Flood et al, Nature 2013; Liu et al, Nature 2012).
ATP-activation and taste stimulation activated the same central cells.
Unless otherwise stated, all sweet stimulations were performed using 2M sucrose.
For most bitter stimulations, we activated bitter-sensing cells by expressing P2X2 under the control of Gr66a-LexA and stimulating with a solution containing 10 mM denatonium, 100 mM caffeine, 100 mM ATP and 20% PEG. This stimulation provided the strongest, most reliable activation, possibly because Gr66a is expressed in all bitter cells whereas most bitter compounds activate a subset of bitter cells (Weiss et al., 2011). For bitter stimulations without P2X2, we used the same solution as above minus ATP. To achieve reliable responses with the solution minus ATP, it was necessary to increase the potassium concentration of the AHL from 5 mM to 12.5 mM (Figure 3A–D only).
Stimulations using only water generated weak responses in taste projections and activated only a very small number of neurons (2–3) in the SEZ. Therefore, we activated water-sensing cells by expressing P2X2 under the control of ppk28-LexA and stimulating with 100 mM ATP in 20% PEG. Stimulation of the proboscis of control flies with 100 mM ATP or 20% PEG did not cause substantial SEZ activity (n=5 brains; mean ± SEM; ATP = 3±1 neurons; PEG = 1±0 neurons; sucrose control = 30±1.1 neurons).
To apply taste solutions to the proboscis, a glass pipette (outer diameter of 1.0 mm and an inner diameter of 0.78 mm) was filled with approximately 2 µL of taste solution using suction generated by a 1 mL syringe. The solution was drawn up the capillary slightly such that the tip of the capillary was empty. The capillary was then placed over the proboscis using a micromanipulator prior to image acquisition. At time point 5 of 20, slight pressure was applied to the syringe to deliver taste solution to the proboscis. Stimulations lasted approximately 2 seconds or the equivalent of 1–2 time points. There were 10 minute intervals between successive stimulations.
For GABAB receptor agonist experiments, SKF97541 was applied at 20 µm final concentration, as previously described (Chu et al., 2014).
Image Acquisition
All experiments were performed using a 3i spinning disk confocal system equipped with a 20× water objective, NA=1, and 1.6× optical zoom with the exception of the higher brain imaging (20× water objective, NA=1, no optical zoom), and the motor neuron fills (40× water objective, NA=1, no optical zoom). The NA of the objective is 1 and the pinhole diameter is 50 µm, leading to an axial resolution of ~2 µm. The point-to-point lateral resolution of the 3i Spinning Disk system is approximately 200 – 250nm. Prior to a stimulation trial, a single stack of 20–23 Z-slices (1.3–1.5 µm) was captured for the green and red channels using 488nm and 561nm lasers respectively. This single stack contained baseline GCaMP fluorescence and nuclear position data and was used as a reference stack. During the stimulation trial, stacks of 20–23 Z-slices (1.3–1.5 µm) were obtained from the green channel at a rate of ~0.5 Hz, for both the SEZ and central brain imaging.
Images were aligned using a rigid body transformation following a regular step gradient descent optimization to minimize the mean square error between two images. As this approach is fundamentally intensity based, slices that showed large changes in fluorescence at individual timepoints occasionally led to misalignment. Automatic alignments that failed or showed poor alignment were aligned manually in all four dimensions. Aligned images could still retain some “jitter” as the brain can exhibit non-rigid movement. However, these deviations were typically no larger than 1–3 pixels. Brains that appeared to undergo significant movement in the z direction were not analyzed.
Images were then aligned with the reference data using custom scripts in MATLAB to determine the nuclear positions of cells during each trial. The onset of stimulation was readily identified by a fluorescence increase in the taste projections. Taste-responsive neurons were identified as cells with fluorescent changes at either the same time point as the taste projection response or at the following time point (from 0-~4 sec post-stimulus) for all images. Images were processed in two ways:
-
1
Circular ROIs with a radius of five pixels were automatically generated in MATLAB by performing circle detection using a Circular Hough Transform on the nuclear data from the reference stack. These ROIs were then used to calculate fluorescence changes for each cell.
Because the cell bodies of many SEZ neurons are positioned on top of dense neuropil (cell bodies are on the fly brain circumference), fluorescence increases from out-of-focus fibers during a stimulation translated into a general fluorescence increase in the plane of the cell bodies. Therefore, using a standard ΔF calculation such as Fluorescencepost-stimulation − Fluorescencepre-stimulation generated artificially high ΔF values for many neurons because of their proximity to responsive neuropil. To correct for this, for each ROI, a circle with a radius of at least 7 pixels was generated. A second, halo-shaped ROI surrounding the first ROI was generated with an inner radius equivalent to the radius of the first ROI and an outer radius 15 pixels larger. At each timepoint, the change in fluorescence of the “halo” ROI was subtracted from the mean fluorescence within the center ROI. This value was used to calculate ΔF/F for each ROI over time. For an initial F value, we used the mean fluorescence intensity of each ROI over the three timepoints prior to stimulation.
For measuring the activity of taste projections, we used the ΔF/F calculation of (Fluorescencepost-stimulation − Fluorescencepre-stimulation)/Fluorescencepre-stimulation (average of 3 previous frames).
-
2
The aligned GCaMP fluorescence data were imported into VAA3D and were manually annotated by marking the center positions of putative responding cells. In some cases, this approach allowed for the identification of neurons that were not otherwise detected because of image processing artifacts and allowed for the elimination of inappropriately labeled neurons that arose because of movement or proximity to taste-responsive fiber tracts. Furthermore, this approach allowed for the rapid identification and elimination of neurons that showed neural activity at multiple time points not associated with taste stimulation. (We found that in approximately 5% of preparations, there is spontaneous rhythmic activity of a few cells (1–2)). The marked cell positions were then exported from VAA3D in the form of “marker” files, imported into MATLAB and converted into circular ROIs for which ΔF/F values were calculated as described above.
A cell was determined to be taste-responsive if its ΔF/F value was greater than 8%. In initial experiments, we performed k-means clustering of all cellular ΔF/F values over multiple stimulations for single brains (6 stimulations for one brain, 3 stimulations for two brains). Clustering placed the boundary between groups at approximately 10% ΔF/F. We also monitored the responses of 1000 ROIs not associated with cell bodies (in neuropil regions) to determine the extent of diffuse signals. Non-cellular ROIs showed a distribution of responses, with a response cloud centered around 0 that extended to the range of 8–10 %. Similar results were obtained from three different brains, justifying an 8% ΔF/F cutoff. No additional responsive cells were identified by increasing the sampling rate 23× (by imaging one Z-section instead of 23 Z-sections at 83ms/section) (Figure S1).
To generate pseudo-colored images of responding cells, ΔF images for every Z-slice at every time point were generated in Matlab. For those time points that contained the peak responses for a given stimulation, the images were masked according to ROI positions such that only cell bodies of responding neurons were visible. These ΔF stacks were either collapsed into a single maximum intensity projection (MIP) image to display the whole SEZ, or collapsed into three shorter MIP images to divide the SEZ into thirds. Then, using FIJI, ΔF images for two stimulations within a given fly were overlaid onto a MIP background image of the original fluorescence data at the time point of maximal response for one of the two stimulations.
To generate summary brain images, we used VAA3D to map cell body positions of responding neurons onto a 3D representation of the raw imaging data. These data could then be rotated in three dimensions to make slight adjustments to correct for variation in brain alignment from preparation to preparation.
Behavior
Female flies, 2–3 days old, were food deprived on a wet kimwipe for 18–24 hrs and mounted in the imaging chamber as described above. The proboscis was allowed to remain free. The head cuticle was then partially dissected and the head was bathed in AHL. Using a pulled glass capillary, the proboscis was stimulated three times with 0.01, 0.1, 0.5, 1, and 2M sucrose solutions and proboscis extensions were recorded. Mixtures of 500mM sucrose and increasing concentrations of bitter were tested serially in the same manner.
Motor Neuron Labeling
For the experiments of Fig. 6A–D to label taste-responsive MNs, tastant stimulations were performed as above using a 3i spinning disk confocal system with a Phasor spatial light modulator (3i, Intelligent Imaging Innovations) coupled to a Ti:Sapphire femtosecond laser (SpectraPhysics). After the stimulation trial, a pre-photoactivation stack was acquired for the green and red channels. A small ROI was drawn in Slidebook (3i, Intelligent Imaging Innovations) and the pharyngeal and labellar nerves were scanned intermittently with 1–2s bouts of 800 nm light, with an interbout delay of 5s, to photoconert C3PA-GFP. This procedure was continued for ~1 hour with a five-minute break to allow for diffusion every ten minutes. At the end of the labeling sequence, a post-photoactivation image stack was taken of the green and red channels.
To generate pseudo-colored images of labeled motor neurons, the pre-photoactivation image was subtracted from the post-photoactivation image and the resulting stack was overlaid onto the ΔF image from the taste stimulation. The labeled neurons were pseudo-colored red and the taste-responsive neurons were pseudo-colored green.
Transgene generation
To generate nSyb-LexA, we cloned an 832 bp fragment from genomic DNA, including the entire R57C10 tile from the FlyLight collection (Pfeiffer et al., 2012), using the primers gtttttaaatttcccaccccttg and gttctagagggttgcgctc. This fragment was recombined into the pBPLexA::p65Uw plasmid (Pfeiffer et al., 2010), and inserted into the attP40 landing site (Markstein et al., 2008).
Supplementary Material
Highlights.
Sweet, bitter, and water sensory neurons activate different cell populations
Some cells respond to more than one modality
Cross-inhibition between different sensory classes governs mixture responses
Bitter and sweet pathways remain segregated in the higher brain
Acknowledgments
The authors thank William E. Allen for writing the initial program for calcium imaging analysis. Dr. Charles Zuker and members of the Scott lab provided advice and comments on the manuscript. This work was supported by a grant from the NIH (NIDCD DC013280) to K.S. K.S. is an HHMI Early Career Scientist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.T.H. designed and performed most experiments, analyzed data and co-wrote the manuscript. B.R.K. performed the C3PA-GFP labeling studies. B.C.M. generated the nSyb-LexA flies. K.S. supervised the project and co-wrote the manuscript.
References
- Accolla R, Bathellier B, Petersen CC, Carleton A. Differential spatial representation of taste modalities in the rat gustatory cortex. J Neurosci. 2007;27:1396–1404. doi: 10.1523/JNEUROSCI.5188-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens MB, Li JM, Orger MB, Robson DN, Schier AF, Engert F, Portugues R. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nature methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- Barretto RP, Gillis-Smith S, Chandrashekar J, Yarmolinsky DA, Schnitzer MJ, Ryba NJ, Zuker CS. The neural representation of taste quality at the periphery. Nature. 2015;517:373–376. doi: 10.1038/nature13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, Waddell S. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492:433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron P, Hiroi M, Ngai J, Scott K. The molecular basis for water taste in Drosophila. Nature. 2010;465:91–95. doi: 10.1038/nature09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton A, Accolla R, Simon SA. Coding in the mammalian gustatory system. Trends in neurosciences. 2010;33:326–334. doi: 10.1016/j.tins.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Gabitto M, Peng Y, Ryba NJ, Zuker CS. A gustotopic map of taste qualities in the mammalian brain. Science (New York, NY. 2011;333:1262–1266. doi: 10.1126/science.1204076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Amrein H. Enhancing Perception of Contaminated Food through Acid-Mediated Modulation of Taste Neuron Responses. Curr Biol. 2014;24:1969–1977. doi: 10.1016/j.cub.2014.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang Q, Wang Z. The amiloride-sensitive epithelial Na+ channel PPK28 is essential for Drosophila gustatory water reception. J Neurosci. 2010;30:6247–6252. doi: 10.1523/JNEUROSCI.0627-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B, Chui V, Mann K, Gordon MD. Presynaptic gain control drives sweet and bitter taste integration in Drosophila. Curr Biol. 2014;24:1978–1984. doi: 10.1016/j.cub.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Foster K, van der Goes van Naters WM, Carlson JR. A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nature neuroscience. 2001;4:1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Gelfand MV, Collins CA, DiAntonio A. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. The Journal of comparative neurology. 2008;508:131–152. doi: 10.1002/cne.21670. [DOI] [PubMed] [Google Scholar]
- Dethier VG. The Hungry Fly. Cambridge, MA: Harvard University Press; 1976. [Google Scholar]
- Diegelmann S, Bate M, Landgraf M. Gateway cloning vectors for the LexA-based binary expression system in Drosophila. Fly. 2008;2(4):236–239. doi: 10.4161/fly.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood TF, Iguchi S, Gorczyca M, White B, Ito K, Yoshihara M. A single pair of interneurons commands the Drosophila feeding motor program. Nature. 2013;499:83–87. doi: 10.1038/nature12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nature methods. 2011;8:253–259. doi: 10.1038/nmeth.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nature reviews. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, et al. A systematic nomenclature for the insect brain. Neuron. 2014;81:755–765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Jeong YT, Shim J, Oh SR, Yoon HI, Kim CH, Moon SJ, Montell C. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 2013;79:725–737. doi: 10.1016/j.neuron.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. 2008;18:1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon CH, Katz DB. The neural processing of taste. BMC neuroscience. 2007;8(Suppl 3):S5. doi: 10.1186/1471-2202-8-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Liman ER, Zhang YV, Montell C. Peripheral Coding of Taste. Neuron. 2014;81:984–1000. doi: 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Placais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, Tanimoto H. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488:512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- Manzo A, Silies M, Gohl DM, Scott K. Motor neurons controlling fluid ingestion in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6307–6312. doi: 10.1073/pnas.1120305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella S, Fischler W, Kong P, Asgarian S, Rueckert E, Scott K. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. 2006;49:285–295. doi: 10.1016/j.neuron.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nature genetics. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Chen Y, Slone J, Amrein H. Identification of a Drosophila glucose receptor using Ca2+ imaging of single chemosensory neurons. PloS one. 2013;8:e56304. doi: 10.1371/journal.pone.0056304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassel DR. Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Progress in neurobiology. 2002;68:1–84. doi: 10.1016/s0301-0082(02)00057-6. [DOI] [PubMed] [Google Scholar]
- Pandey R, Heidmann S, Lehner CF. Epithelial re-organization and dynamics of progression through mitosis in Drosophila separase complex mutants. Journal of cell science. 2005;118:733–742. doi: 10.1242/jcs.01663. [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Developmental cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashekhar KP, Singh RN. Organization of motor neurons innervating the proboscis musculature in Drosophila melanogaster meigen (Diptera: Drosophilidae) International Journal of Insect Morphology and Embryology. 1994;23:225–242. [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- Schoenfeld MA, Neuer G, Tempelmann C, Schussler K, Noesselt T, Hopf JM, Heinze HJ. Functional magnetic resonance tomography correlates of taste perception in the human primary taste cortex. Neuroscience. 2004;127:347–353. doi: 10.1016/j.neuroscience.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Schrodel T, Prevedel R, Aumayr K, Zimmer M, Vaziri A. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nature methods. 2013;10:1013–1020. doi: 10.1038/nmeth.2637. [DOI] [PubMed] [Google Scholar]
- Simon SA, de Araujo IE, Gutierrez R, Nicolelis MA. The neural mechanisms of gustation: a distributed processing code. Nature reviews. 2006;7:890–901. doi: 10.1038/nrn2006. [DOI] [PubMed] [Google Scholar]
- Smith DV, John SJ, Boughter JD. Neuronal cell types and taste quality coding. Physiology & behavior. 2000;69:77–85. doi: 10.1016/s0031-9384(00)00190-6. [DOI] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell. 2012;149:1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Curr Biol. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011;69:258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Yuyama N, Kato T, Kawamura Y. Gustatory responses of cortical neurons in rats. II. Information processing of taste quality. Journal of neurophysiology. 1985;53:1356–1369. doi: 10.1152/jn.1985.53.6.1356. [DOI] [PubMed] [Google Scholar]
- Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. Analysis of functional neuronal connectivity in the Drosophila brain. Journal of neurophysiology. 2012;108:684–696. doi: 10.1152/jn.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.