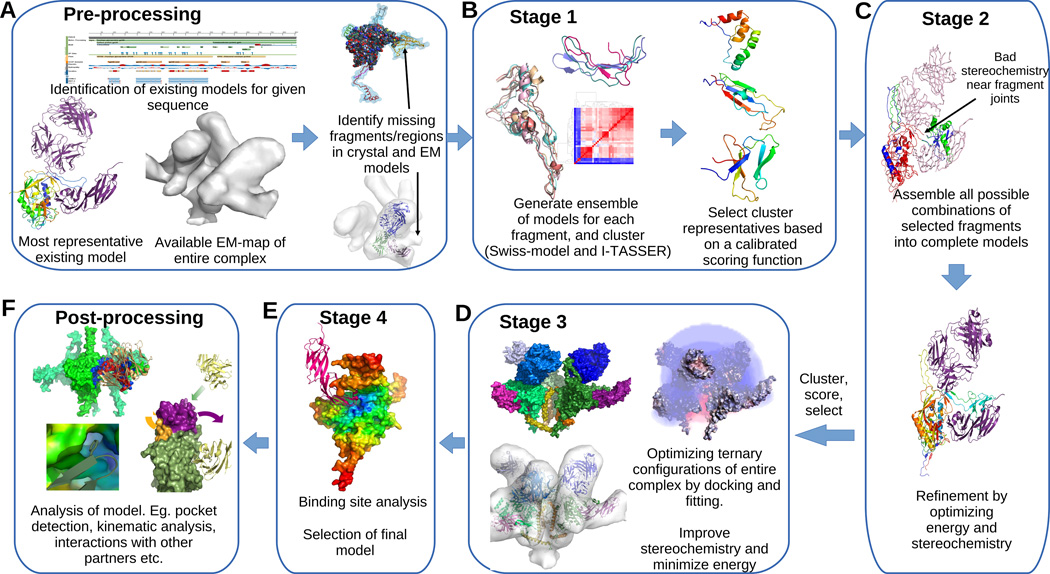
Figure 1. Integrative refinement and validation protocol for modeling proteins with variable domains.
(A) Given a sequence we identify candidate partial crystal structures and EM-maps for the protein and identify the fragments that are missing in the crystal structure and locate corresponding empty regions in the EM. (B) Threading and homology modeling are used to generate an ensemble of candidate models for each fragment. Existing partial crystal structures, known binding interfaces, prior knowledge about residue contacts and stereochemical properties of proteins are used to calibrate a multi-term scoring function, which is used to rank the clusters and select small number of models for each fragment. The same scoring model is used in ranking and selecting models in the remaining steps as well. (C) Fragments are assembled in all possible combinations to generate a large set of complete models, which are then refined iteratively in terms of both the energy and stereochemistry. (D) A small set of refined models are co-optimized with other chains in the complex to improve the ternary interfaces and fitting to the EM-map. A single model (E) is chosen based on the scoring function and binding site analysis. (F) Further analytics are performed to validate the model, to compare with previous models and also to infer new kinematic, energetic, and binding information.