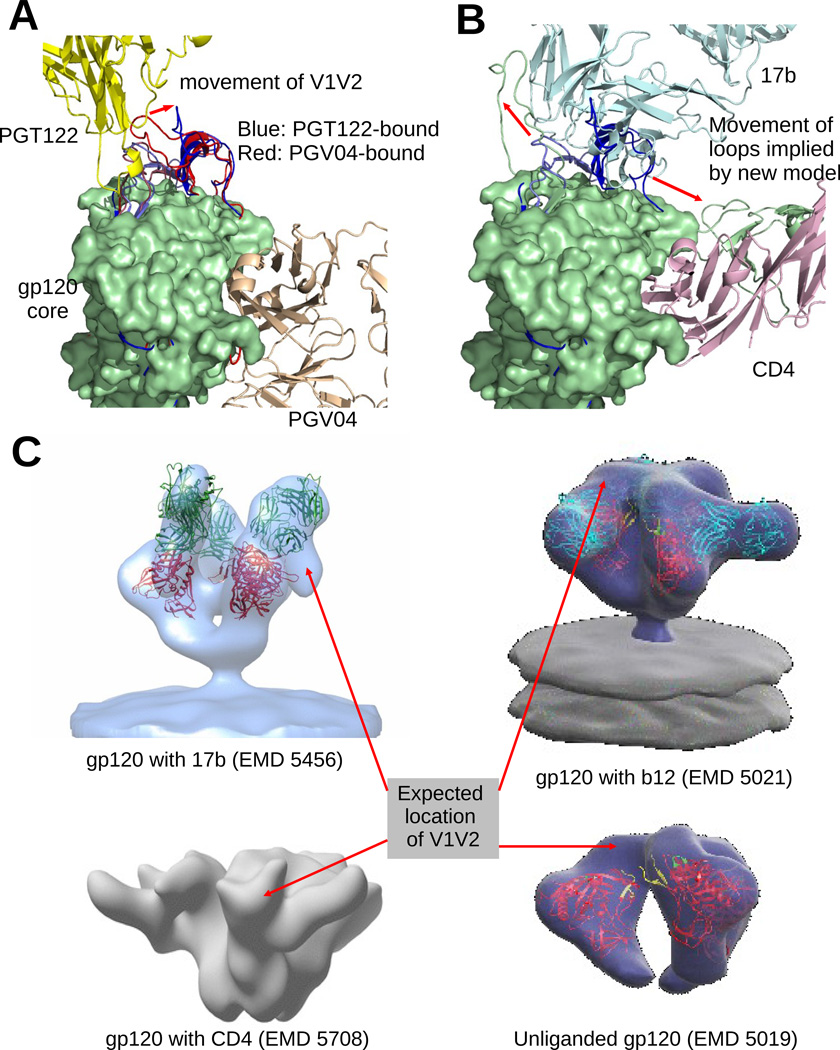
Figure 5. Model suggests probable flexible movement of gp120 variable loops.
(A) Superposition of the gp120 models from 4NCO and 3J5M (blue and red), bound to PGT122 and PGV04 respectively. The V1/V2 and V3 loops occupy the same general location in both cases. But in the PGT122 bound configuration, the V1V2 loop has a noticeable shift to avoid clash with PGT122. In (B), we see that 17b, which binds on the core near the V3 loop has large clash with both V1V2 and V3 loops of 4NCO (blue), and to allow the binding of 17b, both loops must move away to the configuration observed in our model (green). (C) We compared the expected locations of the V1V2 loops in several EM models of gp120. The expected locations are identified by locating large vacant space in the EM map near the V1V2 stem of a fitted atomic model of gp120. EM model of gp120 bound with only 17b (EMD 5456 shows a movement of the V1V2 loop (similar to our reported model) away from the ‘top’ region of gp120. Interestingly, gp120 bound with only soluble CD4 (EMD 5708 also indicates a similar motion. So, the movement of V1V2 is not only a result of repulsion by 17b, but maybe also due to an attraction towards CD4. On the other hand, b12, which also binds at the same site as CD4, does not allow/require V1V2 to move away from its unliganded configuration