Fig 1. ΔLf is modified by SUMOylation.
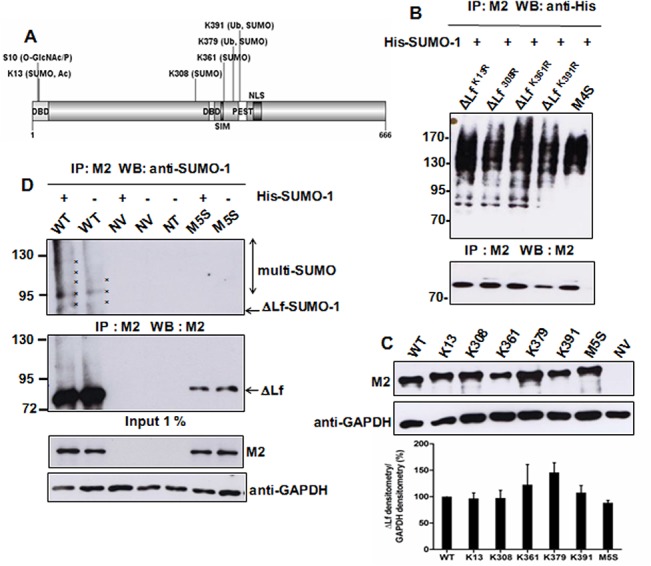
A) Schematic overview of ΔLf showing the NLS and PEST sequences, the two putative DBD and the putative SIM domain. The amino acid residues targeted by post-translational modifications are shown, S10 as the main O-GlcNAc/P site, K379 and K391 as the two ubiquitinated lysines, K13 as a putative acetylation site. B) Mutation of K13, K308, K361 and K391 individual lysine residues did not abolish ΔLf SUMOylation. The first series of ΔLf mutant constructs (ΔLfK13R, ΔLfK308R, ΔLfK361R, ΔLfK391R and the M4S mutant constructs) were co-transfected with the pSG5-His-SUMO-1 (His-SUMO-1) plasmid in HEK-293 cells for 24 h prior to lysis. Lysates were immunoprecipitated with M2 and immunoblotted with anti-His antibodies and M2. The data presented correspond to one representative experiment of two conducted (n = 2). C) Expression of pCMV-3xFLAG-ΔLfWT (WT) and the second series of SUMOylation mutant constructs. WT and the above constructs were transfected for 24 h prior to lysis. Whole cell extract was immunoblotted with either anti-FLAG M2 or anti-GAPDH antibodies. The data presented correspond to one representative experiment of at least seven conducted (n ≥ 7). NV: null vector (pCMV-3xFLAG). The level of expression of each mutant compared to WT is shown in the bar graph beneath the figure (n ≥ 7). D) ΔLf is SUMOylated and M5S is not. WT and the M5S mutant construct were co-transfected with or without the pSG5-His-SUMO-1 (His-SUMO-1) plasmid in HEK-293 cells for 24 h prior to lysis. Lysates were immunoprecipitated with M2 and immunoblotted with anti-SUMO-1 antibodies and M2. Asterisks correspond to SUMO bands (mono-SUMO, 86 kDa; multi-SUMO, 97, 108, 119 kDa). Lysates from HEK-293 cells transfected with a null vector (NV) and from non-transfected (NT) cells were used as negative controls. The data presented correspond to one representative experiment of at least three conducted (n ≥ 3).
