Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-dependent transcription factor. It heterodimerizes with aryl hydrocarbon nuclear translocator, binds to the xenobiotic-responsive element (XRE), and enhances the transcription of genes encoding xenobiotic metabolizing enzymes. AHR also plays important roles in the inhibition of intestinal carcinogenesis and the modulation of gut immunity. It is very important to screen for AHR-activating compounds because those are expected to produce the AHR-mediated physiological functions. Until now, AHR-mediated transcriptional activity represented by the transcriptional activity of CYP1A1 in luciferase assay has been applied as a screening procedure for AHR-activating compounds. However, the AHR-mediated transcriptional activity did not necessarily correspond with the CYP1A1 transcriptional activity. To evaluate AHR-mediated transcriptional activity more specifically, and to screen for AHR-activating compounds, we establish a stable AHR-responsive HepG2 cell line by co-transfection of an AHR expression vector and an AHR-responsive vector (pGL3-XRE) containing a luciferase gene and three tandemly arranged XRE elements into a human hepatoma derived cell line, HepG2. The induction of luciferase activity in the stable AHR-responsive HepG2 cell line by typical AHR activators occurred in time- and concentration-dependent manners. By assessing the AHR target genes CYP1A1, UGT1A1, and ABCG2, an AHR activator-mediated induction was observed at mRNA level. Furthermore, the AHR activator-mediated induction of luciferase activity was positively correlated with the mRNA levels of CYP1A1, UGT1A1, and ABCG2. These findings verified the usefulness of the established stable AHR-responsive HepG2 cell line for the screening of AHR-activating compounds.
Keywords: Aryl hydrocarbon receptor, Luciferase assay, CYP1A1, UGT1A1, ABCG2
Introduction
The aryl hydrocarbon receptor (AHR) is a ligand-dependent transcription factor that is widely represented in human cells and tissues (Pohjanvirta and Tuomisto 1994). AHR belongs to the basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) family and translocates into the nucleus and heterodimerizes with another bHLH/PAS protein, aryl hydrocarbon nuclear translocator (ARNT) (Crews 1998). This AHR/ARNT complex binds to specific enhancer sequences adjacent to target promoters termed xenobiotic responsive elements (XREs) and enhances the transcription of target genes encoding phase 1 metabolic enzymes such as the cytchrome P450 family (CYP) 1A1 and CYP1A2, phase 2 metabolic enzymes such as uridine diphosphate glucuronosyltransferase (UGT) 1A1 and UGT1A6, and phase 3 metabolic enzymes such as the ATP-binding cassette (ABC) transporter, subfamily G, member 2 (ABCG2) (Whitlock 1999; Nakata et al. 2006). Therefore, AHR was originally regarded as a key regulator of xenobiotic metabolism.
Recently, many studies have reported that AHR may have alternate biological roles in addition to its regulation of xenobiotic metabolism. AHR functions as a ligand-dependent E3 ubiquitin ligase of certain nuclear receptors (Ohtake et al. 2007). Kawajiri et al. (2009) reported that AHR played a critical role in the suppression of intestinal carcinogenesis by a previously un-described ligand-dependent mechanism of proteasomal β-catenin degradation and the natural AHR ligands of indole derivatives such as indole-3-carbinol and 3,3-diindolylmethane suppressed intestinal carcinogenesis in an AHR-dependent manner. More recently, there have been some reports that AHR plays an important role in the modulation of the intestinal immune system (Hooper 2011). Some reports have indicated that AHR regulates regulatory T cell (Treg) and T helper 17 (Th17) cell differentiation in gut lamina propria and mesenteric lymph nodes in a ligand-specific manner (Singh et al. 2011). It was also reported that AHR controls the maintenance and differentiation of intestinal lymphoid cells (Kiss et al. 2011; Li et al. 2011). These reports suggest that AHR not only plays a significant role in the regulation of xenobiotic metabolism, but also in the prevention of intestinal carcinogenesis and modulation of gut immunity.
The prototypical AHR ligands are the known environmental contaminants 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), polycyclic aromatic hydrocarbons (PAHs), and halogenated aromatic hydrocarbons (HAHs) (Fernandez-Salguero et al. 1996; Sogawa and Fujii-Kuriyama 1997). Moreover, some dietary compounds have the potential to be AHR ligands. In previous studies, flavonoids (Amakura et al. 2003; Zhang et al. 2003; Wang et al. 2012), indole compounds (Heath-Pagliuso et al. 1998; Degner et al. 2009), and curcumin (Ciolino et al. 1998) were shown to act as natural AHR ligands. An assay system to effectively evaluate AHR-mediated transcriptional activity is needed because a variety of drugs, environmental pollutants, and food nutrients control the physiological processes mediated by AHR.
The aim of this study was to establish a stable cell line for screening of AHR-activating compounds. Until now, AHR-mediated transcriptional activity represented by the transcriptional activity of CYP1A1, whose promoter contains XRE, has been applied as a screening procedure for AHR-activating compounds (Garrison et al. 1996; Natsume et al. 2005; Hamada et al. 2006). However, the AHR-mediated transcriptional activity does not necessarily correspond to the transcriptional activity of CYP1A1 (Aix et al. 1994; Denison et al. 1998).
In order to evaluate the AHR-mediated transcriptional activity more specifically, we established a stable AHR-responsive HepG2 cell line by co-transfection of an AHR expression vector (pcDNA-hAHR) and an AHR-responsive vector (pGL3-XRE) containing three tandemly arranged XRE elements upstream of the luciferase gene into a human hepatoma derived cell line: HepG2. The cell line could be used to evaluate AHR-mediated transcriptional activity by the luciferase assay, but also to study AHR target gene expression at the mRNA level. The utility of the established stable AHR-responsive HepG2 cell line was confirmed in assays of AHR-activating compounds.
Materials and methods
Cell culture
The HepG2 cell line (derived from human hepatic cancer tissue) was obtained from the American Type Culture Collection (ATCCATCC, Manassas, VA, USA). HepG2 cells were grown and maintained at 37 °C in a Dulbecco’s modified Eagle’s medium (DMEM, Wako Pure Chemical Industries, Osaka, Japan) containing 10 % fetal bovine serum (FBS, Asahi Technoglass, Chiba, Japan), 4 mM l-glutamine, 40 U/mL of penicillin, and 40 μg/mL of streptomycin under a humidified atmosphere containing 5 % CO2. For culturing of the stable AHR-responsive HepG2 cells, 500 μg/mL of the neomycin analog G418 sulfate (Nacalai Tesque, Kyoto, Japan) was added to the same medium as that used for culturing the HepG2 cells.
Expression vector and reporter vector construction
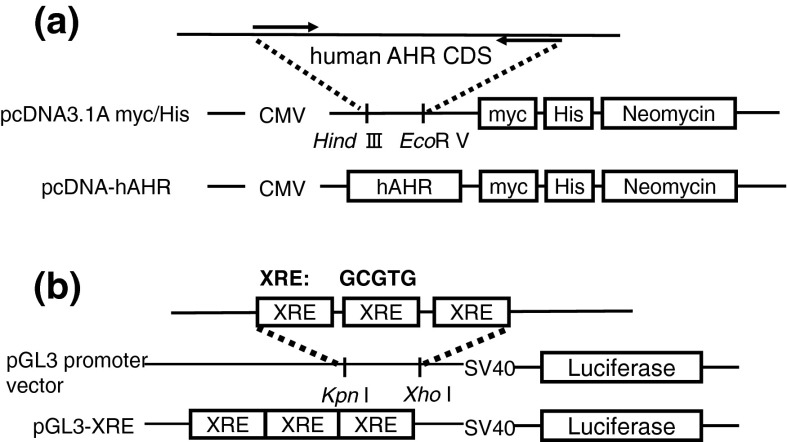
The AHR expression vector (pcDNA-hAHR, Fig. 1a) was constructed as follows: the open reading frame of human AHR was amplified from HepG2 cells by using the primer 5-ATAAGCTTTG GGCACCATGA ACAGC-3, which introduced an EcoRV site, and the primer 5-CCGATATCTC AGGAATCCAC TGGATGTC-3, which introduced a HindIII site. EcoRV/HindIII-digested PCR fragments were ligated into appropriately digested pcDNA3.1/myc-His A (Invitrogen, Carlsbad, CA, USA). The AHR-responsive vector (pGL3-XRE, Fig. 1b) was constructed by ligating the annealed oligonucleotides, which contained three tandemly arranged XREs (GCGTG), into the KpnI/XhoI-digested pGL3 promoter vector (Promega, Madison, WI, USA): sense, 5-CAAGGTACCAGTGCGTGTCAGTGCGTGTCAGTGCGTGTCTCGAGGCT-3, and antisense, 5-AGCCTCGAGACACGCACTGACACGCACTGACACGCACTGGTACCTTG-3.
Fig. 1.
Construction of the AHR expression vector and the AHR-responsive vector. a AHR expression vector (pcDNA-hAHR); b AHR-responsive vector (pGL3-XRE)
Stable integration and selection of G418-resistant colonies
The AHR expression vector and the AHR-responsive vector were stably transfected into HepG2 cells as described previously (Natsume et al. 2005). HepG2 cells were transfected with the expression vector and reporter vector by a lipofectamine technique with lipofectamine and PLUS reagent (Gibco, Gaithersburg, MD, USA). One day before transfection, 2 × 105 cells were plated on 60-mm dishes and allowed to reach 50–60 % confluency prior to replacing the medium in the dishes with a serum-free medium 1 h before transfection. Plasmid DNA (2 μg) and the PLUS reagent (8 μL) were diluted in 250 μL of the serum-free medium and then incubated at room temperature for 15 min. Lipofectamine (12 μL) was diluted in 250 μL of the serum-free medium. The plasmid and lipid dilutions were combined, gently mixed, and then incubated at room temperature for 15 min. Meanwhile, the medium in the dishes was replaced with 2 mL of serum-free medium. A 500-μL portion of the DNA/lipid suspension was added to each dish and gently mixed.
The transfection process was followed by neomycin selection to ensure stable integration of plasmid DNA. After incubating for 3 h, the serum-free medium in the dishes was replaced with normal culture medium. The cells were cultured for 2 weeks in a medium containing 500 μg/mL of G418 disulfate until colonies appeared. The G418-resistant colonies were selected and expanded in a 24-well cell culture plate (Corning-Coster Japan, Tokyo, Japan). Among these, the colonies that showed adequate number of cells to pass to 48 well plates and conduct luciferase assay (more than 50 % confluent/well) were tested for their luciferase activity by treating with 3-methylcholanthlene (3MC, Wako Pure Chemical Industries). The colony that exhibited the greatest increase in luciferase activity by treatment with 3MC was selected as the stable AHR-responsive HepG2 cell line.
Luciferase assay
The luciferase assay was carried out to assess AHR-mediated transcriptional activity as previously described (Hamada et al. 2006). The stable AHR-responsive HepG2 cells were seeded in a 24-well cell culture plate at a density of 1 × 105 cells/well. The 24-well cell culture plate had been pre-coated with Type I–C collagen (Nitta Geratin, Osaka, Japan) for 30 min before the cells were seeded. After an overnight incubation, the medium was replaced with the medium containing test samples. Each test sample was diluted in the cell culture medium at the test concentrations, with the concentration of DMSO never exceeding 0.1 % (v/v) in the cell culture medium. After 24 h of culture, the cells were washed twice with 500 μL of phosphate-buffered saline (PBS), lysed with 100 μL of 1× passive lysis buffer (Promega), the lysate was centrifuged at 15,000×g for 5 min, and the supernatant was collected for the luciferase assay. Luciferase activity was determined by a dual-luciferase reporter assay (Promega) with an LB9507 Lumet luminometer (Berthold Technologies, Bad Wildbad, Germany).
Chemicals
Benzo[a]pyrene (BEN), indigo (IND), 2-(1H-Indol-3-ylcarbonyl)-4-thiazolecarboxylic acid methyl ester (ITE), and 6-formylindolo[3.2-b] carbazole (FICZ) were obtained from Wako Pure Chemical Industries. These chemicals were previously established as candidate AHR ligands (Nguyen and Bradfield 2008). Each phytochemical was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 mM before use.
Isolation of total RNA and real-time PCR
The mRNA expression levels of CYP1A1, UGT1A1, and ABCG2 were evaluated by real-time PCR. These genes were representative drug metabolizing genes those expressions were regulated by AHR (Whitlock 1999; Nakata et al. 2006). The stable AHR-responsive HepG2 cells were seeded onto a pre-coated 24-well cell culture plate at a density of 1 × 105 cells/well. After an overnight incubation, the medium was replaced with a medium containing each test sample. After 24 h of culture, total RNA was extracted from the cells using TRIzol Reagent (Invitrogen). cDNA was prepared from 1 μg of the total RNA. Real-time PCR was performed with SYBR Premix Ex Taq II (TAKARA BIO INC, Otsu, Japan) and 7900HT Fast Real-Time PCR System (Applied Biosystems, Tokyo, Japan). After denaturing at 95 °C for 30 s, PCR was performed for 40 cycles, each of which consisted of denaturing at 95 °C for 5 s, annealing at 60 °C for 30 s, and dissociating at 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. The PCR primers used for CYP1A1 and UGT1A1 were synthesized at Invitrogen, and those used for β-actin and ABCG2 were synthesized at TAKARA BIO INC. The sequence of each primer is shown in Table 1. The mRNA levels of AHR-target genes were normalized to those of β-actin mRNA.
Table 1.
Primers used for Real-Time PCR analysis
| Gene | GenBank ID | Sequence (5′ → 3′) | Tm | |
|---|---|---|---|---|
| β-actin | NM_001101 | Forward | TGGCACCCAGCACAATGAA | 64.7 |
| Reverse | CTAAGTCATAGTCCGCCTAGAAGCA | 62.3 | ||
| CYP1A1 | NM_000499 | Forward | AGATGGTCAAGGAGCACTACA | 69.0 |
| Reverse | CTGGACATTGGCGTTCTCAT | 68.0 | ||
| UGT1A1 | NM_000463 | Forward | TGGCTGTTCCCACTTACTGCAC | 64.6 |
| Reverse | AGGGTCCGTCAGCATGACATC | 64.7 | ||
| ABCG2 | NM_001257386 | Forward | TGCCCAGGACTCAATGCAAC | 64.5 |
| Reverse | TCGATGCCCTGCTTTACCAAATA | 64.7 | ||
Statistical analysis
The data were expressed as the mean ± SD. Time-dependent and dose-dependent effects were analyzed using Tukey multiple comparison test with one-way ANOVA. Statistically significant differences between the control group and each chemically-treated group were determined by Dunnett’s test. The relationship between the induction of AHR-mediated transcriptional activity and AHR-target gene expression was analyzed using the Pearson product-moment correlation coefficient.
Results
Construction of a stable AHR-responsive HepG2 cell line
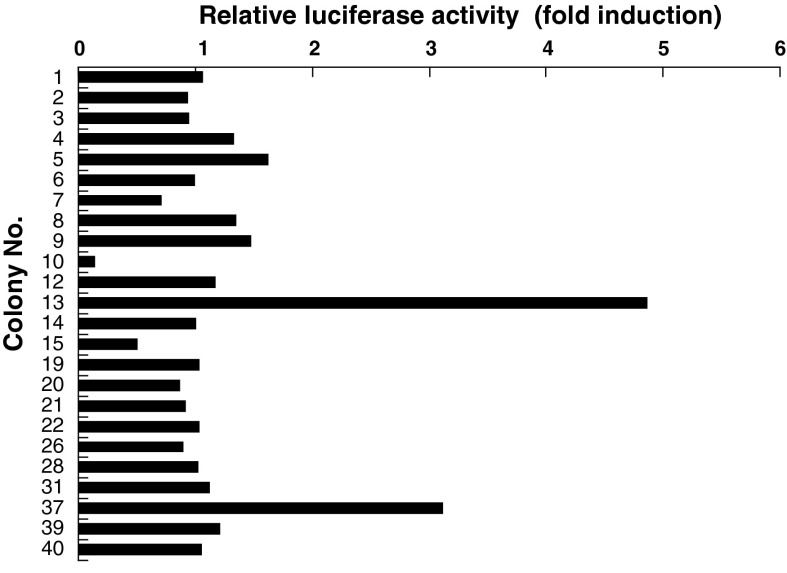
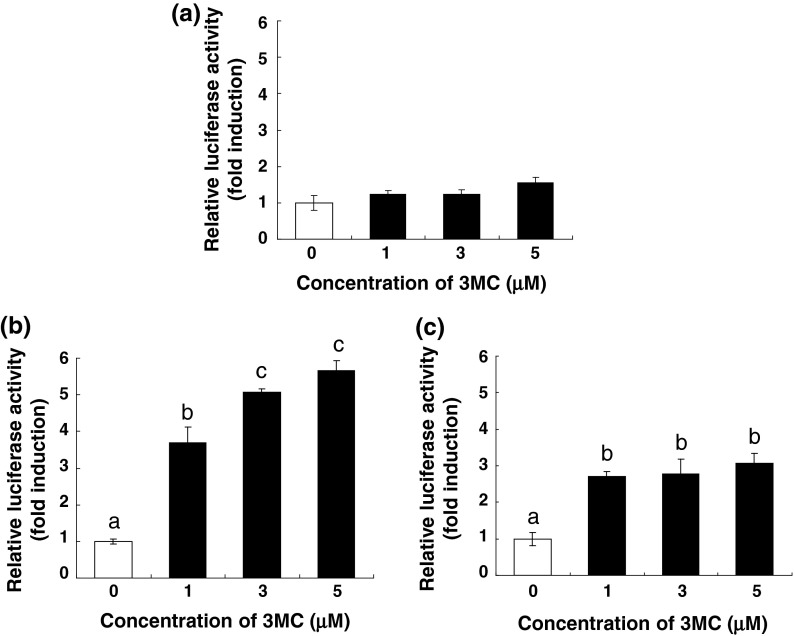
Forty-eight G418-resistant colonies (colony no. 1–48) were selected, and 24 colonies that showed adequate cell growth were tested for their luciferase activity. Three colonies (colony no. 5, 13, and 37) exhibited substantially increased luciferase activity with 3MC treatment, showing more than a 1.5-fold induction (Fig. 2). Furthermore, in colonies 13 and 37, 3MC increased the luciferase activity in a concentration-dependent manner, with the highest increase in luciferase activity being observed in colony 13 (Fig. 3b, c). Thus, colony 13 was selected as the stable AHR-responsive HepG2 cell line.
Fig. 2.
Effect of 3MC on AHR-mediated transcriptional activity in G418-resistant colonies. Each colony was seeded in a collagen-coated 24-well cell culture plate at a density of 1 × 105 cells/well. After an overnight incubation, the cells were incubated with a medium containing 3MC at 5 μM or 0.05 % DMSO (control) for 24 h, and the luciferase assay was performed as described in “Materials and Methods”. The data are represented as a ratio to the control. Each value is the mean (n = 2)
Fig. 3.
Dose dependence of the increase in AHR-mediated transcriptional activity following 3MC treatment in colony 5 (a), 13 (b), and 37 (c). Each colony was seeded in a collagen-coated 24-well cell culture plate at a density of 1 × 105 cells/well. After an overnight incubation, the cells were incubated with a medium containing 3MC at 1, 3, and 5 μM or 0.05 % DMSO (control) for 24 h, and the luciferase assay was performed as described in “Materials and Methods”. The data are represented as a ratio relative to the corresponding controls. Each value is the mean ± SD (n = 3). Dose-dependent effects were analyzed using Tukey multiple comparison test with one- way ANOVA. Points not sharing a common letter are significantly different (p < 0.05)
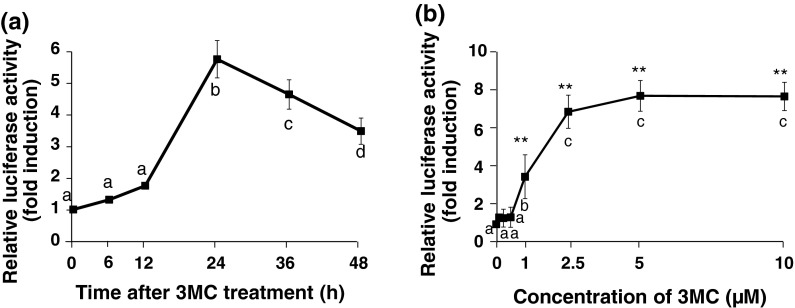
Time- and dose-dependent changes in the AHR-mediated transcriptional activity in stably transfected HepG2 cells after 3MC treatment
The luciferase activity increased in a time-dependent manner up to 24 h after 3MC treatment and gradually decreased thereafter (Fig. 4a). Therefore, the reaction time was fixed at 24 h. The effect of various concentrations of 3MC on the induction of luciferase was further examined. 3MC-mediated increases in luciferase activity occurred in a concentration-dependent manner up to 2.5 μM and were maintained for up to 10 μM (Fig. 4b).
Fig. 4.
Time–course or dose-dependent changes in the AHR-mediated transcriptional activity in stably transfected HepG2 cells after treatment with 3MC. The cells were treated with 3MC (2.5 μM) or with 0.025 % DMSO (control) for the indicated times (a) or were treated with 3MC at the indicated concentrations or 0.1 % DMSO (control) for 24 h (b), and total cell lysates were used for the luciferase assay. Luciferase activity was measured as described in “Materials and Methods”, and the data are represented as a ratio relative to the corresponding controls. Each value is the mean ± SD (n = 3). Time-and dose-dependent effects were analyzed using Tukey multiple comparison test with one-way ANOVA. Points not sharing a common letter are significantly different (p < 0.05)
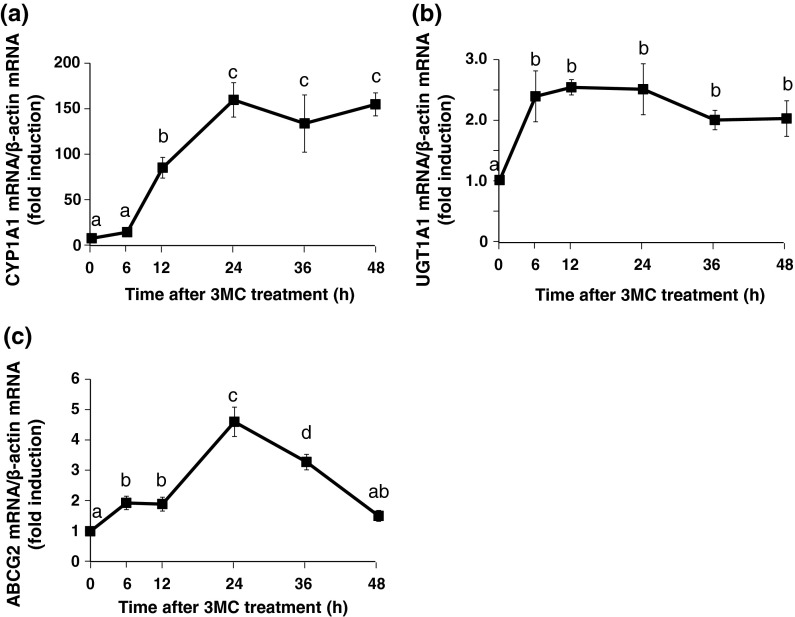
Time–course changes in the mRNA level of the AHR-target genes in stably transfected HepG2 cells after 3MC treatment
CYP1A1, UGT1A1 and ABCG 2 were known as representative target genes of AHR (Whitlock 1999; Nakata et al. 2006). We detected the mRNA expression of these genes to estimate AHR transcriptional activity. The CYP1A1 mRNA levels increased in a time-dependent manner up to 24 h, and this increase was maintained for up to 48 h (Fig. 5a). The expression of UGT1A1 mRNA was maximal at 6 h and was maintained for up to 48 h (Fig. 5b). The ABCG2 mRNA level increased in a time-dependent manner up to 24 h and thereafter gradually decreased (Fig. 5c). Based on these results, the reaction time was fixed at 24 h.
Fig. 5.
Time–course changes in the mRNA level of AHR-target genes in the stably transfected HepG2 cells after treatment with 3MC. The cells were treated with 3MC (2.5 μM) or with 0.025 % DMSO (control) for the indicated times. Total RNA from the cells was used for real-time RT-PCR analysis, and mRNA expression levels of CYP1A1 (a), UGT1A1 (b), and ABCG2 (c) were evaluated. The mRNA levels of AHR-target genes were normalized to those of β-actin mRNA and represented as a ratio relative to the corresponding controls. Each value is the mean ± SD (n = 3). Time-dependent effects were analyzed using Tukey multiple comparison test with one- way ANOVA. Points not sharing a common letter are significantly different (p < 0.05)
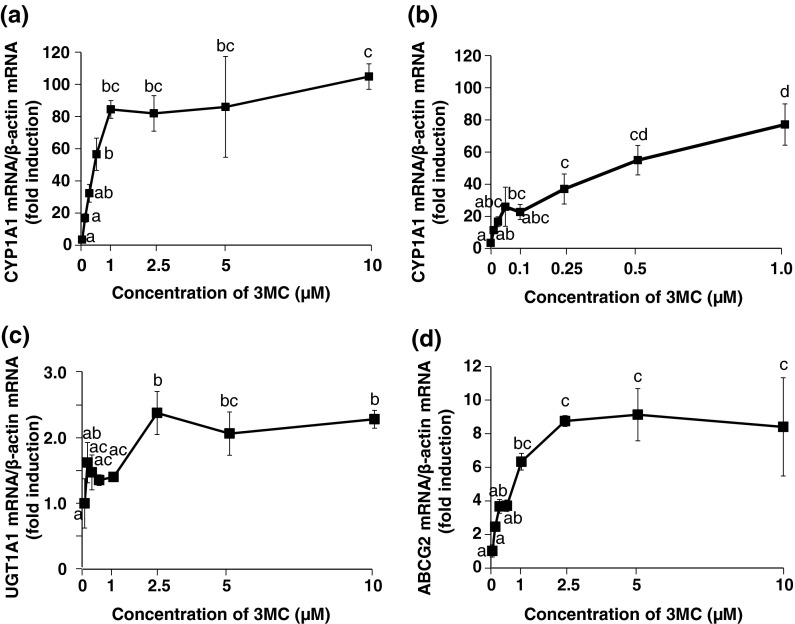
Dose-dependent changes in the mRNA level of AHR-target genes in stably transfected HepG2 cells after treatment with 3MC
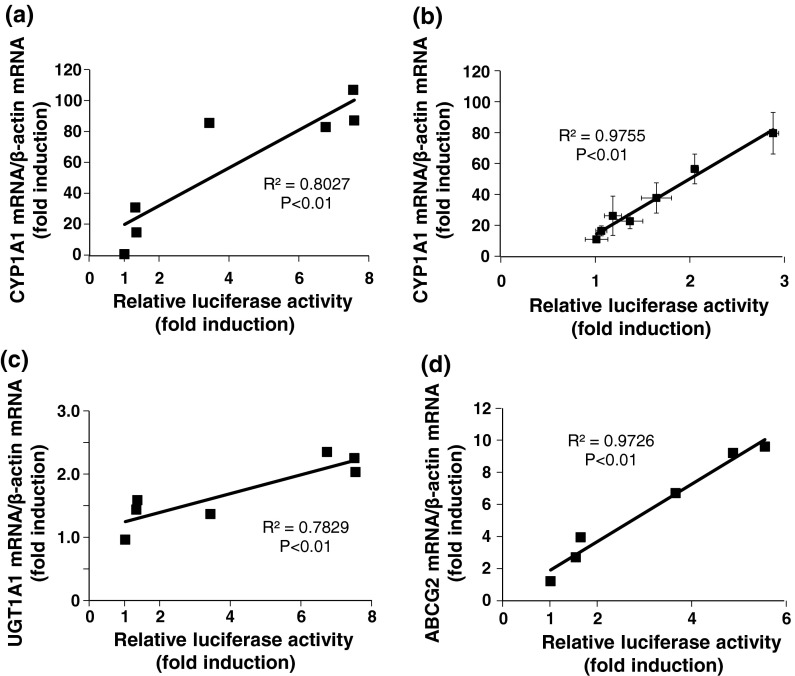
CYP1A1 expression increased in a concentration-dependent manner up to 1 μM (Fig. 6a, b) and maintained for up to 10 μM. Moreover, there was a significant positive correlation between relative luciferase activity and the gene expression of CYP1A1, and the correlation was strong even when the concentration of 3MC was lower than 1 μM (Fig. 7a, b). The 3MC-mediated increase in UGT1A1 mRNA expression was saturated at 2.5-μM 3MC treatment and was maintained for up to 10 μM (Fig. 6c). There was a significant positive correlation between relative luciferase activity and the expression of UGT1A1 (Fig. 7c). ABCG2 expression was increased in a concentration-dependent manner up to 2.5 μM and was maintained for up to 10 μM (Fig. 6d). There was a significant and strong positive correlation between the relative luciferase activity and the gene expression of ABCG2 (Fig. 7d).
Fig. 6.
Dose-dependent changes in the mRNA level of AHR-target genes in the stably transfected HepG2 cells after treatment with 3MC. The cells were treated with 3MC at the indicated concentrations or 0.1 % DMSO (control) for 24 h. Total RNA from the cells was used for real-time RT-PCR analysis, and mRNA expression levels of CYP1A1 (treated with 0–10 μM 3MC) (a), CYP1A1 (treated with 0–1 μM 3MC) (b), UGT1A1 (c), and ABCG2 (d) were evaluated. The mRNA levels of AHR-target genes were normalized to those of β-actin mRNA and represented as a ratio relative to the corresponding controls. Each value is the mean ± SD (n = 3). Dose-dependent effects were analyzed using Tukey multiple comparison test with one- way ANOVA. Points not sharing a common letter are significantly different (p < 0.05)
Fig. 7.
Correlations between luciferase activities and the levels of CYP1A1 (treated with 3MC at 0.1–10 μM) (a), CYP1A1 (treated with 3MC at 0.01–1 μM) (b), UGT1A1 (c), and ABCG2 (d) mRNA after the 3MC-treatment. The relationship between relative luciferase activities and the level of each mRNA was analyzed using the Pearson product-moment correlation coefficient
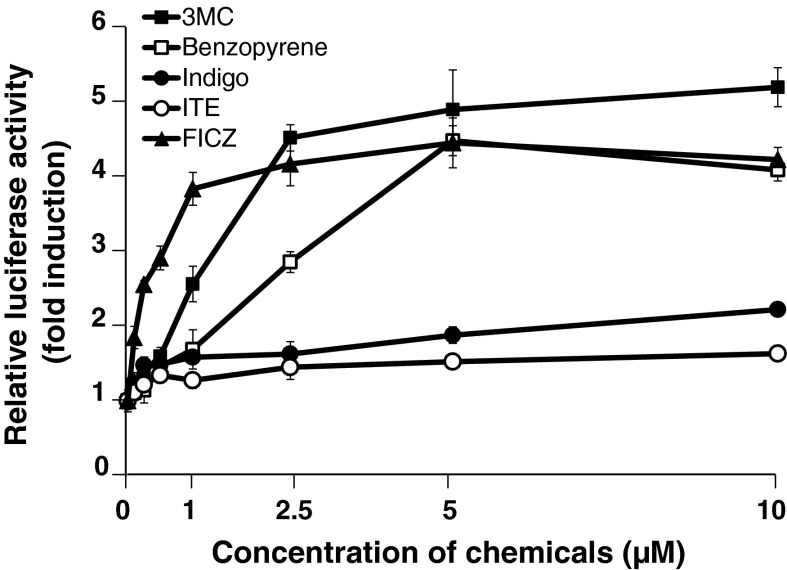
Effects of various chemicals on the induction of AHR-mediated transcriptional activity in the stably transfected HepG2 cells
In this experiment, 3MC, BEN, IND, ITE, and FICZ were used as representative AHR ligands (Nguyen and Bradfield 2008). Treatment of the stable AHR-responsive HepG2 cells with representative AHR ligands for 24 h resulted in significant increases in luciferase activity in a dose-dependent manner (Fig. 8). 3MC, BEN, and FICZ all induced greater luciferase activity than IND and ITE. The AHR-mediated transcriptional activity following FICZ-treatment reached saturation at a lower concentration (1 μM) compared to 3MC and BEN treatment (Fig. 8).
Fig. 8.
Effects of various chemicals on the AHR-mediated transcriptional activity in stably transfected HepG2 cells. The stable AHR-responsive HepG2 cells were treated with each chemical at the indicated concentrations or 0.1 % DMSO (control) for 24 h, and the luciferase assay was performed as described in “Materials and Methods”
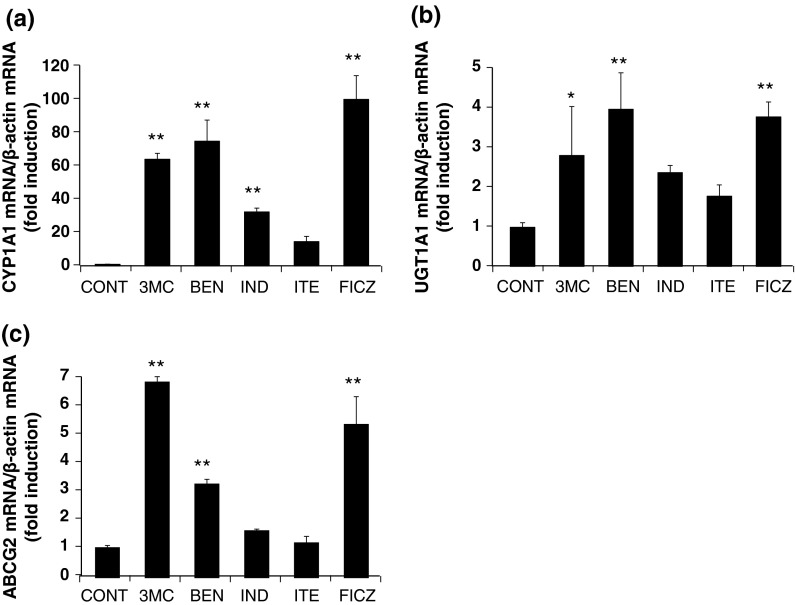
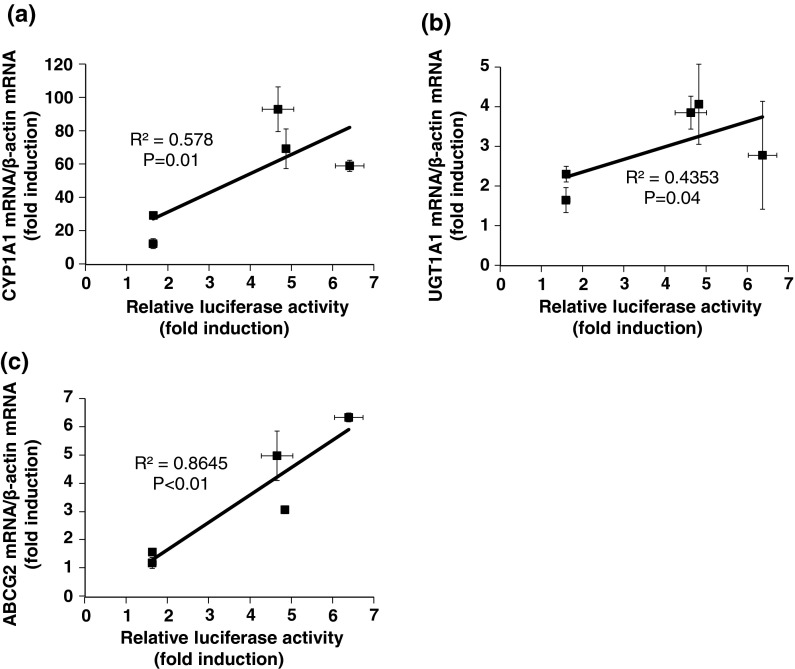
Effects of various chemicals on the induction of AHR-target genes in the stably transfected HepG2 cells 3MC, BEN, and FICZ all induced greater the mRNA expression of AHR-target genes than IND and ITE (Fig. 9a, b, c). This is the same pattern as that obtained for luciferase activity. Relationship analyses revealed a significant and positive correlation between the relative luciferase activity and AHR-target gene expression (Fig. 10a, b, c).
Fig. 9.
Effects of various chemicals on the mRNA level of AHR-target genes in stably transfected HepG2 cells. The stable AHR-responsive HepG2 cells were treated with each chemical (5 μM) or 0.05 % DMSO (control) for 24 h and total RNA from the cells was used for real-time RT-PCR analysis. The levels of CYP1A1 (a), UGT1A1 (b), and ABCG2 (c) mRNA were normalized to that of β-actin mRNA and represented as a ratio relative to the corresponding controls. Significant differences from the corresponding controls assayed were determined by Dunnett’s test; *p < 0.05, **p < 0.01
Fig. 10.
Correlations between relative luciferase activities and the levels of CYP1A1 (a), UGT1A1 (b), and ABCG2 (c) mRNA after chemical treatment. The relationship between relative luciferase activities and the level of each mRNA was analyzed using the Pearson product-moment correlation coefficient
Discussion
In this study, we established the stable AHR-responsive HepG2 cell line by co-transfection of the AHR-responsive vector (pGL3-XRE) and the AHR expression vector (pcDNA-hAHR) into HepG2 cells, and AHR-mediated transcriptional activity in this cell line (based on the luciferase assay) was assessed using 3MC, a representative AHR activator. The time- and concentration-dependent induction of luciferase activity by 3MC was clearly observed in the stable AHR-responsive HepG2 cell line. These effects were comparable to the results obtained using the method for evaluating the CYP1A1 transcriptional activity (Garrison et al. 1996; Fazili et al. 2010). The difference between transcriptional activity of CYP1A1 and AHR-mediated transcriptional activity is not marked when treated with strong AHR-activating compounds such as 3MC. However, CYP1A1 expression is regulated by transcriptional factors other than AHR, including the constitutive androstane receptor (CAR), peroxisome proliferator-activated receptor (PPAR)-α, and the liver X receptor (LXR) (Kaczynski et al. 2002, Sérée et al. 2004, Yoshinari et al. 2010, Shibahara et al. 2011). Therefore, the difference between CYP1A1 transcriptional activity and AHR-mediated transcriptional activity is more considerable when the treatment compounds activate transcriptional factors other than AHR.
Time- and concentration-dependent inductions of AHR target gene transcription by 3MC were observed in the stable AHR-responsive HepG2 cell line, and significant positive correlations between relative luciferase activity and mRNA expression of AHR-target genes were also observed. In previous reports, 3MC induced the gene expression of CYP1A1 (Fazili et al. 2010; Tsuchiya et al. 2003; Stejskalova et al. 2011), UGT1A1 (Smith et al. 2005), and ABCG2 (Tompkins et al. 2010) in many types of cultured tissues and cells. Moreover, it has been reported that 3MC most strongly induces CYP1A1 expression among the three AHR-target genes, CYP1A1, UGT1A1, and ABCG2 (Stejskalova et al. 2011). These reports correspond to the results in this study. There was a significant positive correlation between the induction of AHR-mediated transcriptional activity and CYP1A1, UGT1A1, and ABCG2 mRNA expression (Fig. 7). The correlation between the induction of CYP1A1 mRNA expression and AHR-mediated transcriptional activity is particularly strong in a low concentration range (0.01–1.0 μM). Multiple XREs are located 1,252–892 bp upstream of the CYP1A1 promoter and play an important biological role in regulating CYP1A1 expression (Fujii-Kuriyama et al. 1992; Garrison et al. 1996; Fazili et al. 2010). Conversely, XREs are located approximately 3,424–3,299 bp upstream of the UGT1A1 promoter and 2,357–2,333 bp upstream of the ABCG2 promoter (Yueh et al. 2003; Tompkins et al. 2010). This difference in the location and number of XREs may be responsible for the divergent effects of 3MC on AHR-target gene expression.
Luciferase activity in the stable AHR-responsive HepG2 cell line was increased not only by 3MC, but also by the other known AHR ligands BEN, IND, ITE, and FICZ. This suggests that the stable AHR-responsive HepG2 cell line can be used for the evaluation of AHR-mediated transcriptional activity of AHR-activating compounds. Induction of AHR-mediated transcriptional activity by FICZ plateaued at a lower concentration (1 μM) than the other AHR activators (Fig. 8). The affinity of FICZ for AHR is higher than that of the other AHR agonists used in this study (Nguyen and Bradfield 2008). This characteristic of FICZ may be the reason why the stable AHR-responsive HepG2 cell line responds to low concentrations of FICZ. There was a significant and positive correlation between relative luciferase activity and the gene expression of AHR-target genes (Fig. 10a, b, c). This suggests that the stable AHR-responsive HepG2 cell line can be used for evaluation of AHR-mediated transcriptional activity.
In conclusion, we have established a stable AHR-responsive HepG2 cell line, which is the useful tool for the screening of the AHR-activating compounds. Studies on the induction of AHR-mediated transcriptional activity by drugs or food nutrients are important for understanding the effects of these modulators, because the activation of AHR is associated with multiple physiological functions. Therefore, the established stable AHR-responsive HepG2 cell line will greatly contribute to the understanding of the role of AHR-activating compounds in human health.
Acknowledgments
This work was supported by a Grant-in-Aid for Young Scientists (B-22780115) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.
References
- Aix L, Rey-Grobellet X, Larrieu G, Lesca P, Galtier P. Thiabendazole is an inducer of cytochrome P4501A1 in cultured rabbit hepatocytes. Biochem Biophys Res Commun. 1994;202:1483–1489. doi: 10.1006/bbrc.1994.2098. [DOI] [PubMed] [Google Scholar]
- Amakura Y, Tsutsumi T, Nakamura M, Kitagawa H, Fujino J, Sasaki K, Toyoda M, Yoshida T, Maitani T. Activation of the aryl hydrocarbon receptor by some vegetable constituents determined using in vitro reporter gene assay. Biol Pharm Bull. 2003;26:532–539. doi: 10.1248/bpb.26.532. [DOI] [PubMed] [Google Scholar]
- Ciolino HP, Daschner PJ, Wang TT, Yeh GC. Effect of curcumin on the aryl hydrocarbon receptor and cytochrome P450 1A1 in MCF-7 human breast carcinoma cells. Biochem Pharmacol. 1998;56:197–206. doi: 10.1016/S0006-2952(98)00143-9. [DOI] [PubMed] [Google Scholar]
- Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- Degner SC, Papoutsis AJ, Selmin O, Romagnolo DF. Targeting of aryl hydrocarbon receptor-mediated activation of cyclooxygenase-2 expression by the indole-3-carbinol metabolite 3,3′-diindolylmethane in breast cancer cells. J Nutr. 2009;139:26–32. doi: 10.3945/jn.108.099259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Phelan D, Winter GM, Ziccardi MH. Carbaryl, a carbamate insecticide, is a ligand for the hepatic Ah (dioxin) receptor. Toxicol Appl Pharmacol. 1998;152:406–414. doi: 10.1006/taap.1998.9999. [DOI] [PubMed] [Google Scholar]
- Fazili IS, Jiang W, Wang L, Felix EA, Khatlani T, Coumoul X, Barouki R, Moorthy B. Persistent induction of cytochrome P4501A1 in human hepatoma cells by 3-methylcholanthrene: evidence for sustained transcriptional activation of the CYP1A1 promoter. J Pharmacol Exp Ther. 2010;333:99–109. doi: 10.1124/jpet.109.162222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol Appl Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- Fujii-Kuriyama Y, Imataka H, Sogawa K, Yasumoto K, Kikuchi Y. Regulation of CYP1A1 expression. FASEB J. 1992;6:706–710. doi: 10.1096/fasebj.6.2.1537460. [DOI] [PubMed] [Google Scholar]
- Garrison PM, Tullis K, Aarts JM, Brouwer A, Giesy JP, Denison MS. Species-specific recombinant cell lines as bioassay systems for the detection of 2,3,7,8-tetrachlorodibenzo-p-dioxin-like chemicals. Fundam Appl Toxicol. 1996;30:194–203. doi: 10.1006/faat.1996.0056. [DOI] [PubMed] [Google Scholar]
- Hamada M, Satsu H, Natsume Y, Nishiumi S, Fukuda I, Ashida H, Shimizu M. TCDD-induced CYP1A1 expression, an index of dioxin toxicity, is suppressed by flavonoids permeating the human intestinal Caco-2 cell monolayers. J Agric Food Chem. 2006;54:8891–8898. doi: 10.1021/jf060944t. [DOI] [PubMed] [Google Scholar]
- Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, Denison MS. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry. 1998;37:11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- Hooper LV. You AhR what you eat: linking diet and immunity. Cell. 2011;147:489–491. doi: 10.1016/j.cell.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Kaczynski JA, Conley AA, Fernandez Zapico M, Delgado SM, Zhang JS, Urrutia R. Functional analysis of basic transcription element (BTE)-binding protein (BTEB) 3 and BTEB4, a novel Sp1-like protein, reveals a subfamily of transcriptional repressors for the BTE site of the cytochrome P4501A1 gene promoter. Biochem J. 2002;366:873–882. doi: 10.1042/BJ20020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, Akiyama T, Kurosumi M, Poellinger L, Kato S, Fujii-Kuriyama Y. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci USA. 2009;106:13481–13486. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Nakata K, Tanaka Y, Nakano T, Adachi T, Tanaka H, Kaminuma T, Ishikawa T. Nuclear receptor-mediated transcriptional regulation in Phase I, II, and III xenobiotic metabolizing systems. Drug Metab Pharmacokinet. 2006;21:437–457. doi: 10.2133/dmpk.21.437. [DOI] [PubMed] [Google Scholar]
- Natsume Y, Satsu H, Hamada M, Kitamura K, Okamoto N, Shimizu M. In vitro system for assessing dioxin absorption by intestinal epithelial cells and for preventing this absorption by food substances. Cytotechnology. 2005;47:79–88. doi: 10.1007/s10616-005-3753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, Fujii-Kuriyama Y, Kato S. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- Pohjanvirta R, Tuomisto J. Short-term toxicity of 2,3,7,8-tetrachlorodibenzo -p-dioxinin laboratory animals: effects, mechanisms, and animal models. Pharmacol Rev. 1994;46:483–549. [PubMed] [Google Scholar]
- Sérée E, Villard PH, Pascussi JM, Pineau T, Maurel P, Nguyen QB, Fallone F, Martin PM, Champion S, Lacarelle B, Savouret JF, Barra Y. Evidence for a new human CYP1A1 regulation pathway involving PPAR-alpha and 2 PPRE sites. Gastroenterology. 2004;127:436–445. doi: 10.1053/j.gastro.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Shibahara N, Masunaga Y, Iwano S, Yamazaki H, Kiyotani K, Kamataki T. Human cytochrome P450 1A1 is a novel target gene of liver X receptor α. Drug Metab Pharmacokinet. 2011;26:451–457. doi: 10.2133/dmpk.DMPK-11-RG-030. [DOI] [PubMed] [Google Scholar]
- Singh NP, Singh UP, Singh B, Price RL, Nagarkatti M, Nagarkatti PS. Activation of aryl hydrocarbon receptor (AhR) leads to reciprocal epigenetic regulation of FoxP3 and IL-17 expression and amelioration of experimental colitis. PLoS ONE. 2011;6:e23522. doi: 10.1371/journal.pone.0023522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Graham RA, Krol WL, Silver IS, Negishi M, Wang H, Lecluyse EL. Differential UGT1A1 induction by chrysin in primary human hepatocytes and HepG2 Cells. J Pharmacol Exp Ther. 2005;315:1256–1264. doi: 10.1124/jpet.105.090795. [DOI] [PubMed] [Google Scholar]
- Sogawa K, Fujii-Kuriyama Y. Ah receptor, a novel ligand-activated transcription factor. J Biochem. 1997;122:1075–1079. doi: 10.1093/oxfordjournals.jbchem.a021864. [DOI] [PubMed] [Google Scholar]
- Stejskalova L, Vecerova L, Peréz LM, Vrzal R, Dvorak Z, Nachtigal P, Pavek P. Aryl hydrocarbon receptor and aryl hydrocarbon nuclear translocator expression in human and rat placentas and transcription activity in human trophoblast cultures. Toxicol Sci. 2011;123:26–36. doi: 10.1093/toxsci/kfr150. [DOI] [PubMed] [Google Scholar]
- Tompkins LM, Li H, Li L, Lynch C, Xie Y, Nakanishi T, Ross DD, Wang H. A novel xenobiotic responsive element regulated by aryl hydrocarbon receptor is involved in the induction of BCRP/ABCG2 in LS174T cells. Biochem Pharmacol. 2010;80:1754–1761. doi: 10.1016/j.bcp.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Itoh S, Iwanari M, Yokoi T. Expression of aryl hydrocarbon receptor repressor in normal human tissues and inducibility by polycyclic aromatic hydrocarbons in human tumor-derived cell lines. Toxicol Sci. 2003;72:253–259. doi: 10.1093/toxsci/kfg022. [DOI] [PubMed] [Google Scholar]
- Wang HK, Yeh CH, Iwamoto T, Satsu H, Shimizu M, Totsuka M. Dietary flavonoid naringenin induces regulatory T cells via an aryl hydrocarbon receptor mediated pathway. J Agric Food Chem. 2012;60:2171–2178. doi: 10.1021/jf204625y. [DOI] [PubMed] [Google Scholar]
- Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- Yoshinari K, Yoda N, Toriyabe T, Yamazoe Y. Constitutive androstane receptor transcriptionally activates human CYP1A1 and CYP1A2 genes through a common regulatory element in the 5′-flanking region. Biochem Pharmacol. 2010;79:261–269. doi: 10.1016/j.bcp.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Yueh MF, Huang YH, Hiller A, Chen S, Nguyen N, Tukey RH. Involvement of the xenobiotic response element (XRE) in Ah receptor-mediated induction of human UDP-glucuronosyltransferase 1A1. J Biol Chem. 2003;278:15001–15006. doi: 10.1074/jbc.M300645200. [DOI] [PubMed] [Google Scholar]
- Zhang S, Qin C, Safe SH. Flavonoids as aryl hydrocarbon receptor agonists/antagonists: effects of structure and cell context. Environ Health Perspect. 2003;111:1877–1882. doi: 10.1289/ehp.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]