Figure 3.
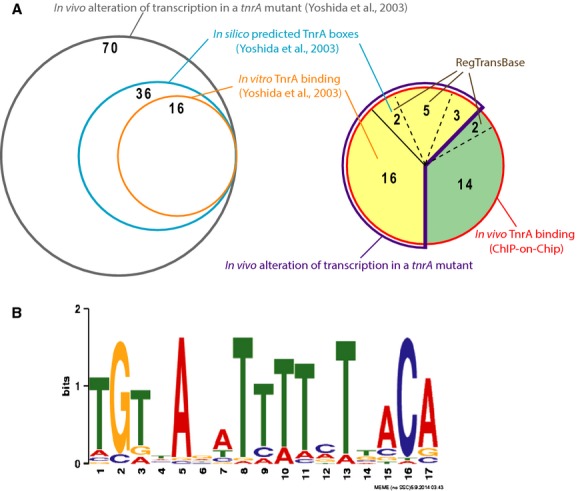
ChIP-on-chip data overlap partly previous studies and allow to generate a new TnrA box consensus. (A) Data obtained in previous studies by transcriptomic and in vitro approaches are presented (left circles) (Yoshida et al. 2003). Forty-two TnrA-binding sites were identified by ChIP-in-chip in this work (red circle, right pie-chart). Twenty-six TnrA-binding sites are associated with promoter regions, whose expression is altered in vivo in a tnrA mutant (yellow area). They belong to the TnrA primary regulon. Sixteen TnrA-binding sites are located in inter- as well as intragenic regions but the role of TnrA as a regulator in the associated regions remains still unknown (green area). They belong to the secondary regulon. Comparison of the ChIP-on-chip data with previous studies is indicated. Sixteen TnrA-binding sites are located in promoter regions previously shown to be directly regulated by TnrA (Wray et al. 2001). In total, nine TnrA-binding sites contain a previously predicted TnrA box (Cipriano et al. 2013). (B) Identification of a new consensus of the TnrA-binding motif. The size of the nucleotide at each position correlates woth its relative prevalence in sequences used as training set in the MEME algorithm (Bailey et al. 2006).
