Figure 3.
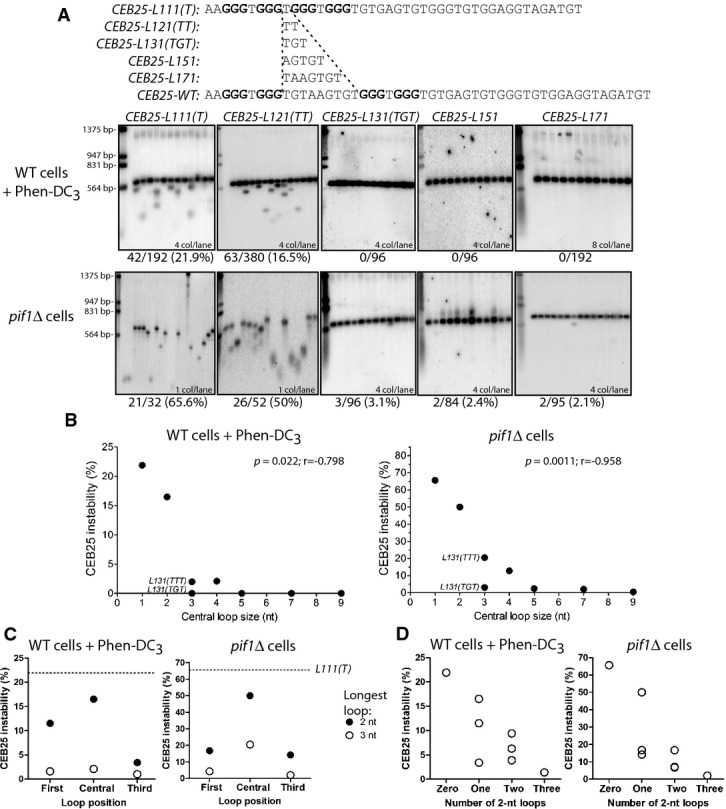
- Southern blot analysis of CEB25 allele variants with shortened central loop length in WT cells treated with Phen-DC3 (top panel) andpif1Δ cells (bottom panel). From left to right: WT strains are ANT1903, ANT1904, ORT7333, ORT7334, and ANT1901;pif1Δ strains are ANT1917, ANT1918, ORT7340, ORT7341, and ANT1902. All the alleles contain 13 motifs. * indicates incompletely digested DNA. Analysis was done as in Fig1B.
- Graphic representation of the instability measurement of central loop length CEB25 variants in WT cells treated with Phen-DC3 (left panel) and inpif1Δ cells (right panel). Instability is inversely correlated to the central loop length in both contexts (two-tailed Spearman correlation test). Alleles bearing sequence modifications other than the central loop (side loops, or intervening sequence) have not been plotted.
- Position effect of a single loop of 2 (filled circles) or 3 nt (open circles) in Phen-DC3-treated WT cells (left panel), and inpif1Δ cells (right panel). Other loops are single residues, and all the nucleotides in loops are thymine. The dotted line denotes the instability of theCEB25-L111(T) allele.
- Effect of the number of 2-nt-long loops (zero, one, two or three) on the CEB25 instability in Phen-DC3-treated WT cells (left panel), and inpif1Δ cells (right panel). Loops that are not 2-nt-long are single residues (consequently the ‘zero’ value corresponds to theCEB25-L111(T) allele). All loops are thymine.
