Figure 6.
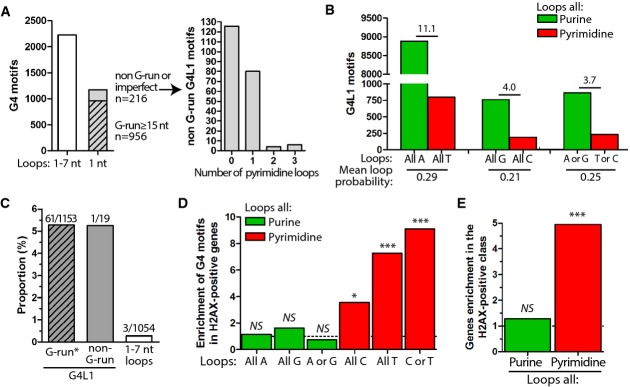
- Analysis of the long- and short-loop G4 motifs in theC. elegans genome. Left panel: number of G4 motifs bearing individual loops up to 7 nt, or single-nucleotide loops only (referred as to ‘G4L1 motifs’). Perfect mono-G-runs (≥ 15 nt, dashed) account for 81.6% (956/1,172) of the G4L1 motifs, imperfect mono-G-runs (with a single loop being different from a G) account for another 16.8% (197/1,172). Only 1.6% (19/1,172) of G4L1 motifs do not belong to the mono-G microsatellite class. Right panel: Pyrimidine loops content among the imperfect and non-G-runs G4L1 motifs (n = 216).
- Composition of the loops of the G4L1 motifs in the human genome. Pairwise comparison between G4L1 motifs bearing exclusively purines (green) and pyrimidines (red) has been performed only for bases with the same probability (e.g., A vs. T), given a mean GC content of 41% for the human genome. We separately analyzed G4L1 motifs bearing identical loops (‘all A’, ‘all T’, etc. as in ourL111 series) from those bearing non-identical loops (e.g., combination of C and T for pyrimidines and A and G for purines), because G4L1 motifs with identical loops are much more prevalent than any of the non-identical G4L1 motifs.
- The 66 non-redundant 100–200-bp deletions mapped in theC. elegans genome upon deletion of thedog-1 helicase (data obtained from Kruisselbrinket al, 2008) are localized at G4L1 motifs. The G4L1 motifs belonging to the G-run* (perfect or imperfect) and the non-G-run classes were equally affected by deletions (5.3% of the sequences in each class), 18-fold more than G4 motifs identified with the least stringent loop length constraint (1–7 nt long). Detailed sequence analysis of these G4 motifs revealed that they still bear short loops (two loops of 1 nt and one loop of 2–4 nt, see Supplementary Fig S7D).
- Fold enrichment of G4L1 motifs by loop composition in γH2AX-positive vs. γH2AX-negative genes following pyridostatin treatment in SV40-infected MRC-5 human fibroblasts (data obtained from Rodriguezet al, 2012). As in (B), we separately analyzed G4L1 motifs bearing identical loops from those bearing non-identical loops. *P < 0.05, ***P < 0.001, NS: non-significant.
- Fold enrichment of genes in the γH2AX-positive vs. γH2AX-negative class depends on the presence of G4L1 motifs bearing pyrimidine loops, but not purine loops. ***P < 0.001, NS: non-significant.
