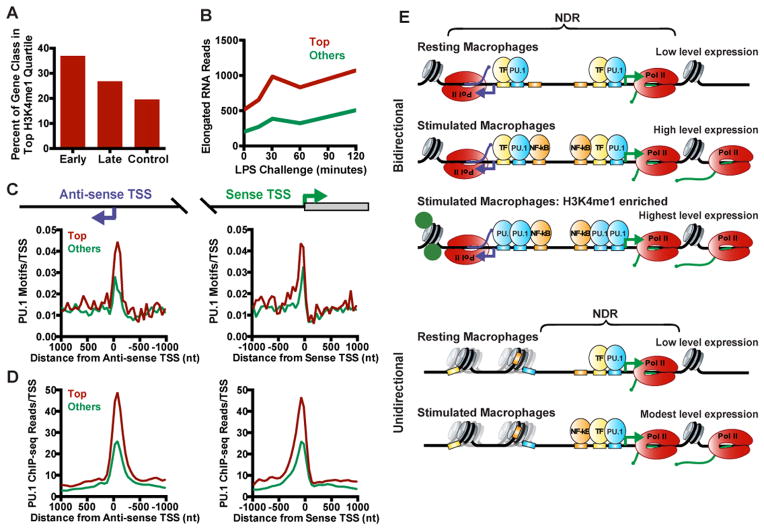
Figure 7. PU.1 Enrichment at Bidirectional TSSs Marks Active and Inducible Genes.
(A) Percentage of Early, Late, and Control genes that fall in the top quartile of H3K4me1 signal around anti-sense TSSs.
(B) Average elongated RNA reads (sense TSS to +250 bp) for genes in the top quartile of H3K4me1 signal, vs. all other bidirectional genes, across a two hour time course of LPS challenge.
(C and D) Average distribution of PU.1 motifs (C) and PU.1 occupancy (D) in 50-mer bins for genes in the top quartile of H3K4me1 vs. all other bidirectional genes, centered on either the anti-sense (left) or sense (right) TSS.
(E) Model for transcriptional activity at bidirectional (top) vs. unidirectional (bottom) promoters. At bidirectional promoters, a lineage-specifying factor such as PU.1 (blue) collaborates with other TFs (yellow) to bind the focused region of TF motifs at coupled sense (green) and anti-sense (purple) TSSs. Through the activity of TFs and the transcription machinery, an NDR is established at both TSSs and spreads between the coupled promoters. This process renders additional transcription factor binding motifs accessible (orange boxes). Following stimulation, signal-dependent transcription factors like NF-κB (orange) preferentially bind motifs in the pre-established NDR enabling high transcriptional output.
At unidirectional promoters, a limited NDR size and absence of well-positioned upstream nucleosome is observed, such that promoter-distal TF motifs are often occluded by chromatin and unavailable for binding.
See also Figure S7.