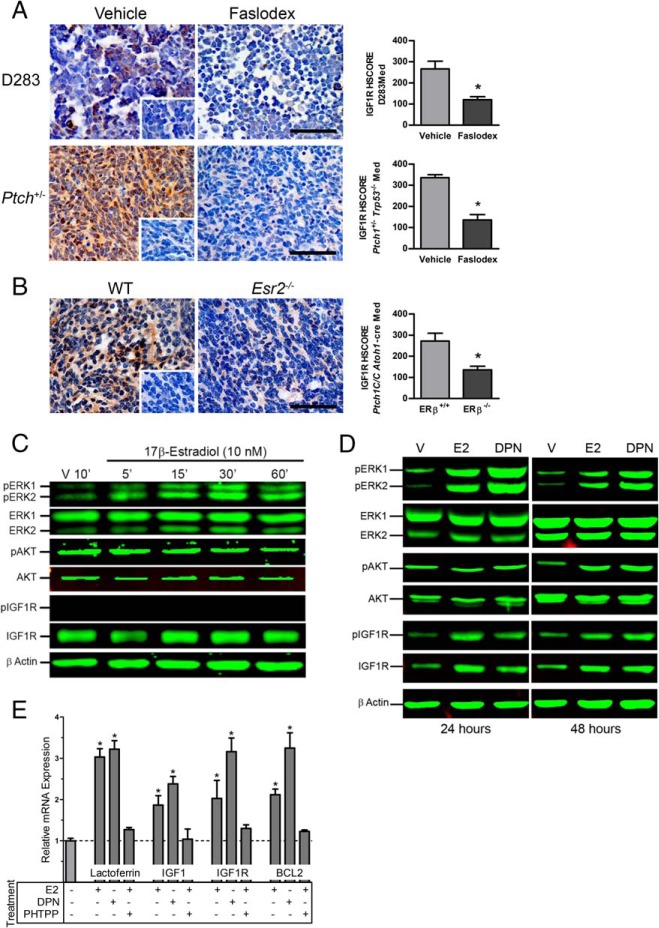
Figure 4.
Immunohistochemical staining of IGF1R in medulloblastoma tumors. Shown are representative micrographs and the results from quantitative analysis of IGF1R immunostaining in D283Med xenografts and Ptch1+/− Trp53−/−tumors from mice treated with vehicle or Faslodex (A) and Esr2+/+ Ptch1C/C Atoh1-cre and Esr−/− Ptch1C/C Atoh1-cre tumors (B). Insets show staining of vehicle treated and Esr2+/+ tumors incubated with normal serum. Quantitative HSCORE results for IGF1R-immunostaing is shown as mean ± SD. Significant differences between mean values for control and experimental group were determined using a Student's t test. For D283Med xenograft groups, n = 5 for vehicle and n = 5 for Faslodex; Ptch1+/− Trp53−/−, n = 4 for vehicle, and n = 4 for Faslodex. For Esr2+/+ Ptch1C/C Atoh1-cre, n = 6 and Esr−/− Ptch1C/C Atoh1-cre, n = 6. Scale bar, 50 μm. Shown are representative results of time-course studies evaluating the temporal profile of ERK1/2, AKT, and IGF1R activation in D283Med cells exposed to 17β-estradiol for up to 60 minutes (C) and with 24- and 48-hour exposures to 17β-estradiol or DPN (D). Total and phospho-specific antigens were detected as indicated and equal loading of proteins were confirmed by simultaneous analysis for β-actin. Treatment times are indicated above each set of panels. Vehicle treated control groups (V) were treated with 0.01% DMSO for indicated times. E, Effects of ER agonists on IGF1, IGF1R, and BCL2 mRNA expression in D283Med cells. Analysis of quantitative RT-PCR analysis of IGF1R and BCL2 mRNA expression detected a 2- to 3-fold increases in the expression of IGF1, IGF1R, and BCL2 after 48 hours of exposure of D283Med cells to E2 or DPN. E2-induced increases in transcript levels were blocked by the ERβ selective antagonist PHTPP. E2 and DPN also induced expression of the positive control estrogen-responsive gene lactoferrin. Significant differences between mean values for expression of each gene target were determined using an ANOVA and Tucky-Kramer posttest from at least three independent experiments.