Abstract
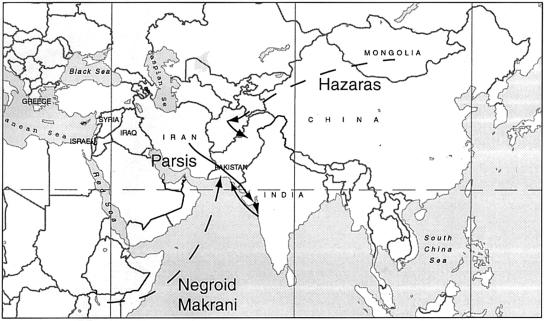
Eighteen binary polymorphisms and 16 multiallelic, short-tandem-repeat (STR) loci from the nonrecombining portion of the human Y chromosome were typed in 718 male subjects belonging to 12 ethnic groups of Pakistan. These identified 11 stable haplogroups and 503 combination binary marker/STR haplotypes. Haplogroup frequencies were generally similar to those in neighboring geographical areas, and the Pakistani populations speaking a language isolate (the Burushos), a Dravidian language (the Brahui), or a Sino-Tibetan language (the Balti) resembled the Indo-European–speaking majority. Nevertheless, median-joining networks of haplotypes revealed considerable substructuring of Y variation within Pakistan, with many populations showing distinct clusters of haplotypes. These patterns can be accounted for by a common pool of Y lineages, with substantial isolation between populations and drift in the smaller ones. Few comparative genetic or historical data are available for most populations, but the results can be compared with oral traditions about origins. The Y data support the well-established origin of the Parsis in Iran, the suggested descent of the Hazaras from Genghis Khan’s army, and the origin of the Negroid Makrani in Africa, but do not support traditions of Tibetan, Syrian, Greek, or Jewish origins for other populations.
Introduction
The earliest evidence of Paleolithic human presence in the Indo-Pakistani subcontinent consists of stone implements found scattered around the Soan River Valley in northern Pakistan (Hussain 1997). Despite the lack of fossil evidence, these tools appear to indicate the presence of hominids in the subcontinent as early as 200,000–400,000 years ago (Wolpert 2000) and thus are likely to have been associated with archaic Homo species. Pakistan lies on the postulated southern coastal route followed by anatomically modern H. sapiens out of Africa, and so may have been inhabited by modern humans as early as 60,000–70,000 years ago. There is evidence of cave dwellers in Pakistan’s northwest frontier, but fossil evidence from the Paleolithic has been fragmentary (Hussain 1997). Evidence has been uncovered at Mehrghar, in southwestern Pakistan, indicating Neolithic settlements from as long ago as 7,000 b.c. (Jarrige 1991), which were followed by the Indus Valley civilizations (including the cities of Harappa and Mohenjodaro) that flourished in the 3d and 2d millennia b.c. (Dales 1991). Around 1500 b.c., the Indo-European–speaking nomadic pastoral tribes from further north—often called the Aryans—crossed the Hindu Kush Mountains into the subcontinent. Subsequent historical events include the invasion of Alexander the Great (327–325 b.c.) and the Arab and Muslim conquest from 711 a.d. onwards (Wolpert 2000).
The present population of Pakistan consists of >150 million individuals (according to current WHO figures) who belong to at least 18 ethnic groups and speak >60 languages (Grimes 1992). Most of these languages are Indo-European, but they also include an isolate, Burushaski; a Dravidian language, Brahui; and a Sino-Tibetan language, Balti. Punjabi-speaking individuals form the majority population of Pakistan, but they represent a complex admixture of ethnic castes and groups (Ibbetson 1883) and are not analyzed here; 12 ethnic groups are included in the present survey. The information available about them is summarized in table 1, together with hypotheses about their origins (Mehdi et al. 1999). Although some of these hypotheses are well-supported (e.g., the origin of the Parsis in Iran), most are based on oral traditions and have not been tested against other sources of evidence.
Table 1.
Pakistani Ethnic Groups Studied
| Ethnic Group | Locationa | Language Family | Census Sizeb | Suggested Origin(s) |
| Baluch | Baluchistan | Indo-European | 4,000,000 | Syria: Aleppo (Quddus 1990) |
| Balti | Eastern Baltistan | Sino-Tibetan | 300,000 | Tibet (Backstrom 1992) |
| Brahui | Baluchistan: Kalat State, Sarawan and Jhalawan regions | Dravidian | 1,500,000 | West Asia (Hughes-Buller 1991) |
| Burusho | Karakorum Mountains: Hunza, Nagar, and Yasin | Language isolate | 50,000–60,000 | Alexander’s army (Biddulph 1977) |
| Hazara | Southern Baluchistan: Quetta and NWFP: Parachinar | Indo-European | NA | Genghis Khan’s soldiers (Bellew 1979) |
| Kalash | NWFP: Hindu Kush Mountains: Bumburet, Rambur, and Birir valleys | Indo-European | 3,000–6,000 | Greeks (Robertson 1896) or “Tsyam,” possibly Syria (Decker 1992) |
| Kashmiri | Kashmir Valley | Indo-European | NA | Jewish (Ahmad 1952) |
| Makrani Baluch | Makran coast | Indo-European | NA | West Asia (Hughes-Buller 1991) |
| Negroid Makrani | Makran coast | Indo-European | NA | Rajput (Quddus 1990); Africa? |
| Parsi | Karachi | Indo-European | A few thousand | Iran, via India (Nanavutty 1997) |
| Pathan | NWFP and Baluchistan | Indo-European | 17,000,000 | Jewish (Ahmad 1952), Greek or Rajput (Bellew 1998; Caroe 1958) |
| Sindhi | Sindh | Indo-European | 15,300,000 | Mixed (Burton 1851) |
NWFP = North West Frontier Province.
NA = not available.
Scanty genetic data are available for these Pakistani ethnic groups. Early studies of the ABO blood groups and classical protein markers did not include all groups and mostly classified them according to their place of residence. A population tree based on 54 classical enzyme markers places the Hazara and Pathans in the West Asian cluster containing the northern Caucasoids (Cavalli-Sforza et al. 1994). In another population tree, based on 47 classical protein polymorphisms, the Pakistani samples form a small subcluster within the Indo-European speakers from India (Cavalli-Sforza et al. 1994).
The Y chromosome provides a unique source of genetic evidence (Tyler-Smith 1999; Jobling and Tyler-Smith 2000). It carries the largest nonrecombining segment in the genome and contains numerous stable binary markers, including base substitutions (see, e.g., Underhill et al. 1997) and retroposon insertions (Hammer 1994; Santos et al. 2000), which can be used in combination with more-rapidly evolving markers, such as microsatellites (see, e.g., Ayub et al. 2000). Consequently, very detailed Y phylogenies can be constructed that allow male-specific aspects of genetic history to be investigated. These are strongly influenced by the small effective population size of the Y chromosome, leading to rapid genetic drift, and by the practice of patrilocality in many societies, leading to high levels of geographical differentiation of Y haplotypes. Notwithstanding the work of Qamar et al. (1999) on the analysis of YAP+ chromosomes (comprising ∼2.6% of the total) and analyses of STR variation (Ayub et al. 2000; Mohyuddin et al. 2001), little work has been carried out on Pakistani Y chromosomes. Therefore, we have now performed an extensive analysis of Pakistani Y lineages, to determine what light they can shed on the origins and genetic history of the subgroups that make up the Pakistani population.
Material and Methods
Samples
The Y chromosomes of 718 unrelated male subjects, belonging to 12 ethnic groups of Pakistan, were analyzed (tables 1 and 2; fig. 1). Informed consent was obtained from all participants in this study. An Epstein Barr virus–transformed lymphoblastoid cell line was established from each individual, and DNA was extracted from these cell lines for analysis.
Table 2.
Diversity, Sample Size (n), and Number of Individuals Belonging to Each Y Haplogroup in 12 Pakistani Ethnic Groups
|
No. of Individuals in Haplogroup |
|||||||||||||
| Population | Diversity | n | 1 | 2 | 3 | 8 | 9 | 10 | 12 | 13 | 21 | 26 | 28 |
| Baluch | .7908 | 59 | 11 | 0 | 17 | 1 | 7 | 0 | 0 | 0 | 5 | 1 | 17 |
| Balti | .7692 | 13 | 2 | 1 | 6 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Brahui | .7516 | 110 | 9 | 11 | 43 | 3 | 31 | 2 | 1 | 1 | 0 | 1 | 8 |
| Burusho | .8065 | 94 | 26 | 7 | 26 | 0 | 7 | 8 | 0 | 0 | 0 | 4 | 16 |
| Hazara | .5573 | 23 | 14 | 1 | 0 | 0 | 1 | 7 | 0 | 0 | 0 | 0 | 0 |
| Kalash | .7558 | 44 | 4 | 17 | 8 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 11 |
| Kashmiri | .6212 | 12 | 3 | 0 | 7 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Makrani Baluch | .8100 | 25 | 6 | 0 | 7 | 1 | 6 | 0 | 0 | 0 | 1 | 0 | 4 |
| Makrani Negroid | .8352 | 33 | 6 | 4 | 10 | 3 | 6 | 0 | 0 | 0 | 1 | 0 | 3 |
| Parsi | .7441 | 90 | 24 | 3 | 7 | 0 | 35 | 0 | 0 | 0 | 5 | 0 | 16 |
| Pathan | .7539 | 93 | 10 | 15 | 42 | 0 | 6 | 0 | 0 | 0 | 2 | 6 | 12 |
| Sindhi | .6937 | 122 | 15 | 11 | 60 | 0 | 25 | 0 | 0 | 0 | 3 | 0 | 8 |
| Overall | .8011 | 718 | 130 | 70 | 233 | 8 | 132 | 17 | 1 | 1 | 17 | 12 | 97 |
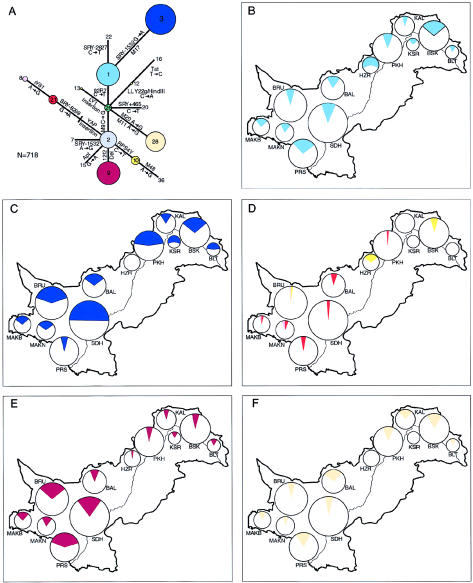
Figure 1.
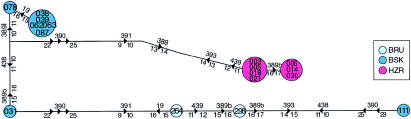
Distribution of Y haplogroups in Pakistan. A, Unrooted maximum-parsimony tree showing Y haplogroups (numbers in circles) and mutations (on lines). Circle area is proportional to frequency in Pakistan. B–F, Frequencies of Y haplogroups in populations sampled. Circles represent populations and are placed in the approximate geographical location sampled; area is proportionate to sample size. BAL = Baluch; BLT = Balti; BRU = Brahui; BSK = Burusho; HZR = Hazara; KAL = Kalash; KSR = Kashmiri; MAKB = Makrani Baluch; MAKN = Makrani Negroid; PKH = Pathan; PRS = Parsi; and SDH = Sindhi. Haplogroup color codes are as in A. B, Haplogroup 1. C, Haplogroup 3. D, Haplogroups 21, 8, and 10. E, Haplogroup 9. F, Haplogroup 28.
Binary Polymorphism Typing
We typed 15 SNPs, an Alu insertion (Hammer 1994; Hammer and Horai 1995), a LINE1 insertion (Santos et al. 2000), and the 12f2 deletion (Casanova et al. 1985). The base substitutions were: 92R7 C→T (Mathias et al. 1994); M9 C→G (Underhill et al. 1997); SRY-2627 C→T (Bianchi et al. 1997); SRY-1532 A→G→A (Whitfield et al. 1995; Kwok et al. 1996; Santos et al. 1999b); sY81 (DYS271) A→G (Seielstad et al. 1994); SRY-8299 G→A (Santos et al. 1999a); Apt G→A (Pandya et al. 1998); SRY +465 C→T (Shinka et al. 1999); LLY22g C→A and Tat T→C transition (Zerjal et al. 1997). In addition, the M17 marker (Underhill et al. 1997) was typed, by use of the primers GTGGTTGCTGGTTGTTACGT and AGCTGACCACAAACTGATGTAGA followed by AflIII digestion of the PCR product; the ancestral allele was not digested. The M20 marker (Underhill et al. 1997) was genotyped, by use of the primers CACACAACAAGGCACCATC and GATTGGGTGTCTTCAGTGCT followed by SspI digestion; the A→G mutation destroys the site at position 118 in the 413-bp product. M11 (Underhill et al. 1997) was typed, using the primers TTCATCACAAGGAGCATAAACAA and CCCTCCCTCTCTCCTTGTATTCTACC followed by digestion with MspI. The 215-bp product was digested to 193-bp and 22-bp fragments in the derived allele. The RPS4Y C→T mutation (Bergen et al. 1999) was detected by BslI restriction digestion of a 528-bp PCR product obtained by use of the primers CCACAGAGATGGTGTGGGTA and GAGTGGGAGGGACTGTGAGA. The ancestral C allele contains two sites, and the derived T allele contains one. M48 (Underhill et al. 1997), A→G, was typed by allele-specific PCR using the discriminating primers TGACAATTAGGATTAAGAATATTATA and TGACAATTAGGATTAAGAATATTATG and the common primer AAAATTCCAAGTTTCAGTGTCACATA to generate specific 145-bp products. The set of Y binary marker alleles carried by a single individual will be referred to as “the Y haplogroup.”
Of the 718 samples, 717 fell into haplogroups expected on the basis of the known phylogeny, but one Pathan sample (PKH134) failed to amplify at the SRY –1532 and M17 loci. He was assigned to haplogroup 3 on the basis of alternative SRY –1532 primers (details on request) and his STR profile.
Y-STR Typing
Five trinucleotide-repeat polymorphisms (DYS388, DYS392, DYS425, DYS426, and DYS436), ten tetranucleotide-repeat polymorphisms (DYS19, DYS389I, DYS389b, DYS390, DYS391, DYS393, DYS434, DYS435, DYS437, and DYS439) and one pentanucleotide microsatellite (DYS438) were typed in all Y chromosomes. Three multiplex PCR reactions were performed for all Y-STRs, in a10-μl final reaction volume containing 20 ng genomic DNA, as described elsewhere (Thomas et al. 1999; Ayub et al. 2000). PCR products were run on an ABI 377 sequencer. ABIGS350 TAMRA was used as the internal lane standard. The GENESCAN and GENOTYPER software packages were used to collect the data and to analyze fragment sizes. Y-STR alleles were named according to the number of repeat units they contain.The number of repeat units was established through the use of sequenced reference DNA samples. Allele lengths for DYS389b were obtained by subtraction of the DYS389II allele length from DYS389I.
Y-STR duplications were found at several loci. DYS393 was duplicated in PKH165 (13 and 15) and DYS437 was duplicated in SDH181 (8 and 9). A more complex pattern was found in DYS425, where two to four alleles were found in 36 individuals from haplogroups 8, 9, 13, and 21.
Data Analysis
Principal-components analysis was carried out on haplogroup frequencies by use of the ViSta (Visual Statistics) system software, version 5.0.2 (Young and Bann 1996). For graphic representation, the first and second principal components were plotted by the Microsoft Office Suite Excel Package on Windows 2000. Biallelic polymorphism data for various world populations used in the analysis were obtained from Hammer et al. (2001). Admixture was estimated by use of three different measures: Long’s weighted least-squares (WLS) measure (Long 1991); mR, a least-squares estimator (Roberts and Hiorns 1965); and mρ (Helgason et al. 2000).
Analysis of molecular variance (AMOVA) was carried out by use of the Arlequin package (Schneider et al. 1997). AMOVA measures the proportions of mutational divergence found within and between populations, respectively. Although much of the variation at the rapidly mutating microsatellite loci is expected to have been produced in the different Pakistani subpopulations, the unique mutation events at the binary loci are much older and have not occurred in the context of the subdivision of the Pakistani population. We devised the following strategy to exploit the maximum amount of relevant mutational information from the Y-chromosome haplotypes. STR variation within haplogroups was used to calculate population pairwise ΦST values for each individual haplogroup. For each population pair, a weighted mean ΦST was calculated, where the value obtained for each haplogroup was weighted according to the proportion of pairwise comparisons involving that haplogroup. In the absence of a particular haplogroup from one population, A, of the pair A and B, ΦST was set to 1, and the number of pairwise comparisons was taken as the number of chromosomes carrying that haplogroup in B. Values of ΦST based on STRs alone or on STRs plus binary markers, with binary markers given a 10-fold higher weighting, were calculated for comparison. In all of these analyses, the distance matrix used consisted of the number of steps by which each pair of haplotypes differed. Mantel tests for the significance of correlations between ΦST values were carried out in Arlequin, and multidimensional scaling (MDS) plots were constructed by use of the SPSS version 7.0 software package.
Median-joining networks were constructed by Network 2.0b (Bandelt et al. 1999). A weighting scheme with a five-fold range was used in the construction of the networks. The weights assigned were specific for each haplogroup and took into account the Y-STR variation across the haplogroup in the whole Pakistani population. The following weights were used: variance 0-0.09, weight 5; variance 0.1-0.19, weight 4; variance 0.2-0.49, weight 3; variance 0.5-0.99, weight of 2; and variance ⩾1.00, weight 1. Despite this, the network for haplogroup 1 contained many high dimensional cubes and was resolved by applying the reduced median and median joining network methods sequentially. The reduced median algorithm (Bandelt et al. 1995) was used to generate a *.rmf file and the median joining network method was applied to this file.
BATWING (Wilson and Balding 1998), Bayesian Analysis of Trees With Internal Node Generation, was used to estimate the time to the most recent common ancestor (TMRCA) of a set of chromosomes. This program uses a Markov chain Monte Carlo procedure to generate phylogenetic trees and associated parameter values consistent with input data (a set of Y haplotypes) and genetic and demographic models. The genetic model assumes single-step mutations of the STRs and the demographic model chosen was exponential growth from an initially constant-sized population, with or without subdivision in different runs of the program. All 16 STR loci were used; locus-specific mutation rate prior probabilities based on the data of Kayser et al. (Kayser et al. 2000) were constructed for the loci available as gamma distributions of the form gamma(a, b) where a = (1 + number of mutations observed by Kayser et al.), and b = (1 + number of meioses). For loci not investigated by Kayser et al., the distribution gamma (1,416) was used, which has a mean of 0.0024. A generation time of 25 years was assumed. Thus the 95% confidence intervals given take into account uncertainty in mutation rate, population growth and (where appropriate) subdivision, but not generation time.
Results
Y-Chromosome Binary Polymorphisms
The 18 binary markers used identify 20 haplogroups in worldwide populations (fig. 1A), but only 11 were found in Pakistan, and 5 accounted for 92% of the sample (fig. 1 and table 2). Haplogroups 1 and 9 were present in all Pakistani populations examined, haplogroup 3 was present in all except the Hazaras, and haplogroup 28 was present in all except the Hazaras and the Kashmiris. Southwestern populations show higher frequencies of hg 9 and the YAP+ haplogroups 21 and 8 than northeastern populations (figs. 1D–E), but, overall, little geographical clustering of haplogroup frequencies is apparent within the country.
Principal-Components Analysis
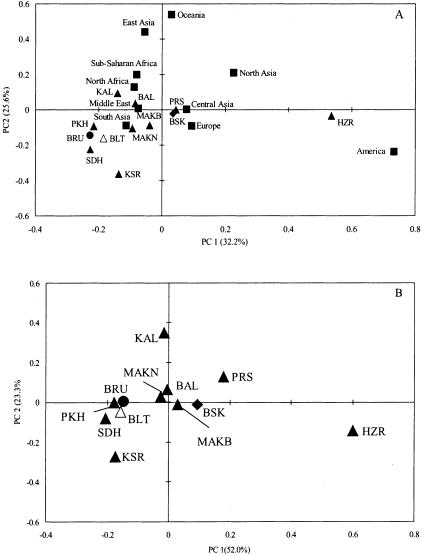
We wished to compare the Pakistani Y haplogroup data with data from populations from the rest of the world. No suitable data set was available for the entire set of 18 markers, but the data of Hammer et al. (2001) allowed all but 5 to be used, because the same or phylogenetically equivalent markers were reported. The principal-components analysis (fig. 2A) shows some differences from the original analysis of Hammer et al., the main one being the lesser separation of the African populations. This is due, to a large extent, to the subset of markers used, which does not include many of the Africa-specific ones. Most Pakistani populations cluster with South Asian and Middle Eastern populations, and are close to Northern African, Central Asian and European populations, thus showing a general similarity with geographically close populations. The one exception is the Hazara, who are quite distinct. A similar analysis of the Pakistani populations alone, using all of the binary markers (fig. 2B), confirms the difference between the Hazaras and the other populations and also more clearly shows the distinctness of the Kalash and the Parsis. It is striking that the language isolate–speaking Burusho and the Dravidian-speaking Brahuis do not stand out in these analyses.
Figure 2.
Principal-components analysis of Y haplogroup frequencies. A, World data using 13 markers. B, Pakistani data using 18 markers. Population codes are as in figure 1. World data are shown as squares. Within Pakistan, Indo-European speakers are indicated by blackened triangles, Sino-Tibetan speakers by an unblackened triangle, Dravidian speakers by a circle, and the language-isolate Burusho by a diamond.
Admixture Estimates
Hypotheses about population origins (table 1) can be considered as quantitative questions about admixture. For example, to test the possibility that the Baluch Y chromosomes have a Syrian origin, we can ask what proportion of the Baluch Ys are derived from Syria and what proportion are from Pakistan (considered to be the Pakistani sample minus the Baluch). Data on suggested source populations were taken from the literature and three measures of admixture were calculated. The three estimates gave broadly consistent results, with small systematic differences: typically mρ > mR > Long’s WLS for the estimated contribution from the external source population (table 3). These results provide evidence for an external contribution to the Hazaras, Kalash, Negroid Makrani, and Parsis but not to the other populations.
Table 3.
Admixture Estimates
|
Admixture Estimate |
|||
| Pakistaniand SourcePopulations | Long's WLS | mR | mρ |
| Baluch: | |||
| Syriaa | −.08 | −.1 | 0 |
| Pakistan | 1.08 | 1.1 | 1 |
| Balti: | |||
| Tibetb | −.06 | −.11 | 0 |
| Pakistan | 1.06 | 1.11 | 1 |
| Burusho: | |||
| Greecec | −.29 | −.22 | 0 |
| Pakistan | 1.29 | 1.22 | 1 |
| Hazara: | |||
| Mongoliab | .67 | .52 | .41 |
| Pakistan | .33 | .48 | .59 |
| Kalash: | |||
| Greecec | .4 | .32 | .23 |
| Pakistan | .6 | .68 | .77 |
| Kashmiri: | |||
| Jewsa | −.46 | −.36 | 0 |
| Pakistan | 1.46 | 1.36 | 1 |
| Negroid Makrani: | |||
| Sub-Saharan Africab | .12 | .12 | .13 |
| Pakistan | .88 | .88 | .88 |
| Pathan: | |||
| Greecec | −.03 | −.16 | 0 |
| Pakistan | 1.03 | 1.16 | 1 |
| Jewsa | −.22 | −.55 | 0 |
| Pakistan | 1.22 | 1.55 | 1 |
| Parsis: | |||
| Irand | 1.21 | 1.06 | 1 |
| Pakistan | −.21 | −.06 | 0 |
Y-Chromosome STR Polymorphisms
Y-STR polymorphisms were studied to obtain a more detailed view of Y variation, among the different Pakistani ethnic groups, that would be less biased by the marker-ascertainment procedure. The diversity of Y-STR haplotypes (table 4) was lowest for the Hazara (0.893) as suggested by previous analyses (Ayub et al. 2000).
Table 4.
STR Variation within the 12 Ethnic Groups from Pakistan[Note]
|
Value for Population |
||||||||||||
| Variable | BAL | BLT | BRU | BSK | HZR | KAL | KSR | MAKB | MAKN | PRS | PKH | SDH |
| n | 59 | 13 | 110 | 94 | 23 | 44 | 12 | 25 | 33 | 90 | 93 | 122 |
| ka | 48 | 12 | 85 | 63 | 11 | 26 | 8 | 24 | 30 | 60 | 72 | 101 |
| HDb±SE | .988 ± .027 | .987 ± .069 | .973 ± .024 | .987 ± .024 | .893 ± .040 | .952 ± .030 | .894 ± .067 | .997 ± .036 | .992 ± .033 | .974 ± .025 | .984 ± .028 | .995 ± .023 |
Note.— The populations are represented by their three- or four-letter codes, as defined in figure 1.
k = Number of lineages.
HD = Haplotype diversity.
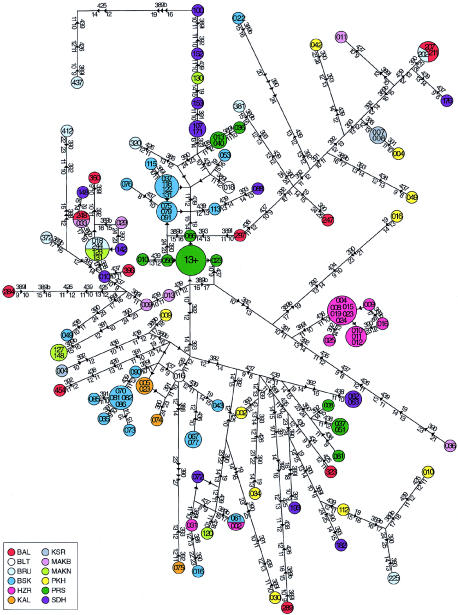
The 16 Y-STRs defined 502 Y haplotypes, the vast majority being observed in single individuals. The remaining haplotypes were shared by 2–18 individuals (details are given in the online-only supplementary table). In all cases but one, the chromosomes sharing a haplotype belonged to the same haplogroup (hence, 503 combination haplotypes) and, in most cases, the individuals sharing a haplotype belonged to the same population (table 5).
Online-Only Supplementary Table.
Y-Chromosomal DNA Haplotypes in Pakistani Populations
|
Repeat Count at Each Y-STR Locus |
No. of Individuals in Populationa |
|||||||||||||||||||||||||||
| Haplogroup and Haplotype | DYS19 | DYS388 | DYS389I | DYS389b | DYS390 | DYS391 | DYS392 | DYS393 | DYS425 | DYS426 | DYS434 | DYS435 | DYS436 | DYS437 | DYS438 | DYS439 | BAL | BLT | BRU | BSK | HZR | KAL | KSR | MAKB | MAKN | PRS | PKH | SDH |
| 1: | ||||||||||||||||||||||||||||
| 1 | 14 | 12 | 10 | 15 | 25 | 11 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 13 | 2 | 1 | ||||||||||
| 2 | 14 | 12 | 10 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 11 | 11 | 10 | 1 | |||||||||||
| 3 | 14 | 12 | 10 | 16 | 23 | 10 | 14 | 13 | 11 | 12 | 8 | 12 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 4 | 14 | 12 | 10 | 16 | 24 | 10 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 12 | 1 | |||||||||||
| 5 | 14 | 12 | 9 | 15 | 23 | 10 | 10 | 14 | 13 | 11 | 9 | 11 | 12 | 9 | 11 | 11 | 1 | |||||||||||
| 6 | 14 | 13 | 10 | 17 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | 1 | ||||||||||
| 7 | 14 | 13 | 9 | 16 | 24 | 10 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 8 | 15 | 12 | 10 | 16 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 12 | 1 | |||||||||||
| 9 | 15 | 12 | 11 | 16 | 23 | 11 | 11 | 13 | 12 | 13 | 9 | 11 | 12 | 8 | 12 | 12 | 1 | |||||||||||
| 10 | 15 | 12 | 9 | 15 | 23 | 11 | 16 | 13 | 13 | 11 | 9 | 11 | 12 | 8 | 12 | 12 | 1 | |||||||||||
| 11 | 14 | 12 | 11 | 16 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 12 | 15 | 12 | 12 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 11 | 1 | |||||||||||
| 13 | 14 | 12 | 10 | 15 | 25 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 13 | 1 | |||||||||||
| 14 | 14 | 12 | 10 | 17 | 23 | 10 | 10 | 12 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 15 | 14 | 12 | 9 | 19 | 23 | 11 | 11 | 13 | 14 | 11 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 16 | 14 | 13 | 10 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 2 | 2 | ||||||||||
| 17 | 14 | 15 | 10 | 16 | 22 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 18 | 15 | 13 | 11 | 16 | 24 | 10 | 10 | 14 | 12 | 11 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 19 | 16 | 12 | 9 | 17 | 25 | 10 | 14 | 13 | 12 | 12 | 9 | 12 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 20 | 13 | 12 | 11 | 17 | 22 | 10 | 15 | 13 | 10 | 12 | 9 | 11 | 12 | 9 | 11 | 11 | 1 | 1 | ||||||||||
| 21 | 13 | 13 | 10 | 16 | 23 | 10 | 15 | 13 | 10 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 22 | 14 | 12 | 10 | 15 | 20 | 10 | 13 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 13 | 1 | |||||||||||
| 23 | 14 | 12 | 10 | 16 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 4 | |||||||||||
| 24 | 14 | 12 | 10 | 16 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 25 | 14 | 12 | 10 | 16 | 23 | 11 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 26 | 14 | 12 | 10 | 16 | 23 | 9 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 27 | 14 | 12 | 10 | 17 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 10 | 1 | |||||||||||
| 28 | 14 | 12 | 11 | 16 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 10 | 9 | 11 | 11 | 2 | |||||||||||
| 29 | 15 | 12 | 10 | 15 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 11 | 1 | |||||||||||
| 30 | 15 | 12 | 10 | 16 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 31 | 15 | 12 | 11 | 15 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 11 | 4 | |||||||||||
| 32 | 15 | 12 | 11 | 15 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 12 | 3 | |||||||||||
| 33 | 15 | 12 | 11 | 15 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 34 | 15 | 12 | 11 | 16 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 11 | 11 | 1 | |||||||||||
| 35 | 15 | 12 | 11 | 16 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 13 | 1 | |||||||||||
| 36 | 16 | 12 | 11 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 11 | 1 | |||||||||||
| 37 | 13 | 12 | 10 | 16 | 22 | 10 | 15 | 13 | 10 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 38 | 14 | 12 | 10 | 17 | 22 | 11 | 13 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 39 | 14 | 12 | 10 | 17 | 23 | 11 | 13 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 12 | 6 | |||||||||||
| 40 | 14 | 12 | 10 | 17 | 23 | 11 | 14 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 41 | 14 | 12 | 10 | 18 | 22 | 11 | 13 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 42 | 14 | 12 | 10 | 18 | 23 | 11 | 13 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 12 | 3 | |||||||||||
| 43 | 14 | 12 | 11 | 16 | 23 | 11 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 12 | 2 | |||||||||||
| 44 | 14 | 12 | 11 | 16 | 24 | 11 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 45 | 14 | 14 | 11 | 16 | 27 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 46 | 14 | 12 | 11 | 17 | 23 | 10 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 47 | 14 | 13 | 10 | 16 | 24 | 11 | 13 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 12 | 2 | |||||||||||
| 48 | 13 | 12 | 11 | 17 | 21 | 11 | 14 | 14 | 12 | 11 | 9 | 12 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 49 | 14 | 12 | 11 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 1 | |||||||||||
| 50 | 14 | 12 | 11 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 11 | 1 | |||||||||||
| 51 | 14 | 12 | 12 | 15 | 24 | 11 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 10 | 12 | 12 | 1 | |||||||||||
| 52 | 14 | 13 | 10 | 15 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 53 | 13 | 12 | 11 | 16 | 22 | 10 | 15 | 13 | 10 | 12 | 9 | 11 | 12 | 9 | 10 | 11 | 1 | |||||||||||
| 54 | 14 | 12 | 11 | 15 | 24 | 12 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 2 | |||||||||||
| 55 | 14 | 12 | 11 | 16 | 23 | 11 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 56 | 14 | 12 | 11 | 15 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 12 | 1 | |||||||||||
| 57 | 14 | 12 | 11 | 15 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 13 | 1 | |||||||||||
| 58 | 14 | 12 | 11 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 12 | 13 | |||||||||||
| 59 | 14 | 12 | 11 | 16 | 24 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 12 | 1 | |||||||||||
| 60 | 14 | 12 | 11 | 17 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 12 | 1 | |||||||||||
| 61 | 15 | 12 | 10 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 12 | 10 | 1 | |||||||||||
| 62 | 15 | 12 | 11 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 12 | 10 | 2 | |||||||||||
| 63 | 15 | 12 | 11 | 16 | 24 | 10 | 14 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 12 | 11 | 2 | |||||||||||
| 64 | 15 | 12 | 11 | 16 | 24 | 10 | 14 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 65 | 15 | 12 | 11 | 16 | 24 | 10 | 15 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 66 | 13 | 12 | 11 | 16 | 22 | 10 | 14 | 14 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 13 | 1 | |||||||||||
| 67 | 13 | 12 | 11 | 19 | 21 | 10 | 14 | 14 | 12 | 12 | 10 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 68 | 14 | 12 | 10 | 16 | 23 | 11 | 13 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 14 | 1 | |||||||||||
| 69 | 14 | 12 | 10 | 17 | 24 | 11 | 12 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 13 | 12 | 1 | |||||||||||
| 70 | 14 | 12 | 11 | 16 | 23 | 10 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 13 | 1 | |||||||||||
| 71 | 14 | 12 | 11 | 18 | 23 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 72 | 14 | 13 | 10 | 16 | 24 | 10 | 13 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 12 | 1 | |||||||||||
| 73 | 15 | 12 | 9 | 16 | 24 | 10 | 14 | 14 | 12 | 12 | 9 | 12 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 74 | 16 | 12 | 10 | 16 | 25 | 11 | 15 | 13 | 10 | 12 | 9 | 11 | 12 | 9 | 11 | 13 | 1 | |||||||||||
| 75 | 16 | 12 | 9 | 18 | 24 | 10 | 14 | 14 | 12 | 11 | 9 | 12 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 76 | 13 | 12 | 11 | 16 | 22 | 10 | 15 | 13 | 10 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 77 | 14 | 12 | 10 | 15 | 24 | 11 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 12 | 14 | 1 | |||||||||||
| 78 | 14 | 12 | 10 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 79 | 14 | 12 | 10 | 16 | 23 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 11 | 1 | |||||||||||
| 80 | 14 | 12 | 10 | 18 | 24 | 9 | 14 | 13 | 12 | 12 | 9 | 12 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 81 | 14 | 12 | 11 | 16 | 23 | 10 | 16 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 11 | 2 | |||||||||||
| 82 | 14 | 12 | 11 | 16 | 23 | 11 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 11 | 1 | |||||||||||
| 83 | 14 | 13 | 10 | 15 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 84 | 15 | 12 | 11 | 16 | 23 | 10 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 2 | |||||||||||
| 85 | 15 | 12 | 11 | 16 | 23 | 11 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 86 | 15 | 12 | 11 | 16 | 24 | 10 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 14 | 1 | |||||||||||
| 87 | 15 | 12 | 9 | 16 | 25 | 10 | 14 | 15 | 12 | 12 | 9 | 12 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 88 | 15 | 13 | 10 | 17 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 2: | ||||||||||||||||||||||||||||
| 89 | 17 | 12 | 11 | 16 | 22 | 7 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 90 | 14 | 12 | 11 | 18 | 23 | 11 | 12 | 12 | 12 | 12 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 91 | 14 | 12 | 11 | 18 | 24 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 11 | 9 | 10 | 11 | 1 | |||||||||||
| 92 | 14 | 12 | 9 | 17 | 23 | 10 | 12 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 93 | 14 | 12 | 9 | 17 | 23 | 11 | 12 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 94 | 14 | 12 | 9 | 19 | 22 | 10 | 11 | 13 | 14 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 95 | 14 | 13 | 9 | 17 | 23 | 10 | 10 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 96 | 15 | 12 | 10 | 17 | 22 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 97 | 15 | 12 | 9 | 17 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 98 | 15 | 12 | 9 | 18 | 22 | 10 | 11 | 13 | 12 | 11 | 9 | 12 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 99 | 15 | 12 | 9 | 19 | 22 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 100 | 17 | 12 | 10 | 17 | 22 | 10 | 11 | 12 | 12 | 13 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 101 | 14 | 12 | 9 | 16 | 23 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 102 | 15 | 12 | 11 | 16 | 22 | 11 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 103 | 15 | 12 | 11 | 16 | 22 | 11 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 104 | 15 | 12 | 12 | 13 | 22 | 11 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 105 | 15 | 13 | 10 | 17 | 22 | 10 | 11 | 12 | 12 | 11 | 9 | 12 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 106 | 15 | 15 | 9 | 16 | 24 | 10 | 11 | 12 | 10 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 107 | 16 | 12 | 10 | 17 | 23 | 11 | 11 | 13 | 11 | 11 | 9 | 11 | 11 | 10 | 10 | 11 | 1 | |||||||||||
| 108 | 14 | 13 | 10 | 16 | 22 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | 1 | |||||||||||
| 109 | 14 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 110 | 14 | 12 | 9 | 16 | 23 | 10 | 15 | 12 | 16 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 111 | 15 | 12 | 10 | 16 | 22 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 2 | |||||||||||
| 112 | 15 | 12 | 10 | 16 | 23 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 3 | |||||||||||
| 113 | 15 | 12 | 11 | 16 | 23 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 114 | 15 | 12 | 9 | 16 | 23 | 10 | 11 | 14 | 14 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 115 | 15 | 13 | 10 | 16 | 22 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 116 | 15 | 13 | 10 | 16 | 23 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 117 | 16 | 12 | 9 | 16 | 23 | 10 | 11 | 14 | 14 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 6 | |||||||||||
| 118 | 14 | 13 | 9 | 17 | 22 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 119 | 15 | 12 | 10 | 18 | 21 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 120 | 15 | 12 | 11 | 14 | 22 | 11 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 13 | 2 | |||||||||||
| 121 | 14 | 12 | 9 | 17 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 122 | 15 | 12 | 11 | 16 | 22 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 123 | 17 | 12 | 11 | 16 | 22 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 124 | 14 | 12 | 11 | 16 | 22 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 125 | 14 | 12 | 11 | 17 | 22 | 10 | 11 | 13 | 12 | 11 | 12 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 126 | 15 | 12 | 10 | 16 | 23 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 13 | 1 | |||||||||||
| 127 | 15 | 12 | 11 | 19 | 21 | 11 | 11 | 14 | 14 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 128 | 16 | 12 | 10 | 15 | 24 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 8 | 11 | 1 | |||||||||||
| 129 | 16 | 12 | 10 | 17 | 22 | 11 | 11 | 13 | 11 | 11 | 9 | 11 | 11 | 10 | 10 | 12 | 1 | |||||||||||
| 130 | 16 | 12 | 10 | 17 | 23 | 11 | 11 | 13 | 11 | 11 | 9 | 11 | 11 | 10 | 10 | 12 | 2 | |||||||||||
| 131 | 16 | 12 | 10 | 17 | 24 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 11 | 11 | 10 | 11 | 1 | |||||||||||
| 132 | 16 | 12 | 11 | 17 | 23 | 10 | 11 | 13 | 11 | 11 | 9 | 11 | 11 | 10 | 10 | 11 | 1 | |||||||||||
| 133 | 16 | 12 | 11 | 18 | 23 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 11 | 10 | 10 | 11 | 1 | |||||||||||
| 134 | 16 | 12 | 9 | 17 | 23 | 11 | 11 | 13 | 11 | 11 | 9 | 11 | 11 | 10 | 10 | 12 | 1 | |||||||||||
| 135 | 16 | 12 | 9 | 18 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 11 | 10 | 10 | 11 | 1 | |||||||||||
| 136 | 17 | 12 | 10 | 17 | 23 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 11 | 10 | 10 | 11 | 1 | |||||||||||
| 137 | 16 | 12 | 9 | 17 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 14 | 1 | |||||||||||
| 138 | 14 | 12 | 11 | 16 | 23 | 10 | 11 | 12 | 12 | 12 | 9 | 13 | 12 | 8 | 10 | 10 | 1 | |||||||||||
| 139 | 14 | 12 | 11 | 16 | 24 | 10 | 11 | 14 | 12 | 11 | 10 | 11 | 11 | 8 | 10 | 12 | 1 | |||||||||||
| 140 | 14 | 12 | 9 | 17 | 22 | 10 | 12 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 141 | 15 | 12 | 10 | 16 | 21 | 10 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 142 | 15 | 12 | 10 | 17 | 21 | 10 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 143 | 15 | 12 | 10 | 18 | 21 | 10 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 144 | 15 | 12 | 11 | 17 | 23 | 10 | 10 | 12 | 10 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 145 | 15 | 12 | 11 | 18 | 23 | 10 | 11 | 13 | 12 | 10 | 9 | 11 | 13 | 8 | 10 | 10 | 1 | |||||||||||
| 146 | 15 | 12 | 9 | 18 | 22 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 147 | 16 | 12 | 11 | 14 | 22 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 148 | 18 | 12 | 11 | 14 | 22 | 10 | 11 | 12 | 13 | 11 | 8 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 3: | ||||||||||||||||||||||||||||
| 149 | 15 | 12 | 10 | 17 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 2 | |||||||||||
| 150 | 15 | 12 | 11 | 15 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 8 | 11 | 1 | |||||||||||
| 151 | 15 | 12 | 11 | 16 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 7 | 7 | 11 | 1 | |||||||||||
| 152 | 15 | 12 | 11 | 17 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | ||||||||||
| 153 | 15 | 12 | 11 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 8 | 11 | 12 | 7 | 10 | 10 | 1 | |||||||||||
| 154 | 15 | 12 | 11 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | 1 | 1 | ||||||||
| 155 | 15 | 12 | 11 | 18 | 26 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 156 | 15 | 12 | 11 | 19 | 24 | 10 | 12 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 157 | 16 | 12 | 10 | 17 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | 10 | 1 | ||||||||
| 158 | 16 | 12 | 10 | 18 | 25 | 10 | 11 | 13 | 12 | 13 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 159 | 16 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 1 | |||||||||||
| 160 | 16 | 12 | 9 | 17 | 25 | 11 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 4 | 1 | 1 | 3 | ||||||||
| 161 | 17 | 12 | 10 | 17 | 25 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 162 | 15 | 12 | 9 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 163 | 16 | 12 | 10 | 16 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | ||||||||||
| 164 | 16 | 12 | 10 | 17 | 24 | 10 | 13 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 165 | 16 | 12 | 10 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 2 | 1 | 2 | 1 | ||||||||
| 166 | 17 | 12 | 10 | 18 | 25 | 10 | 11 | 13 | 11 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 167 | 13 | 12 | 10 | 16 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 168 | 14 | 10 | 11 | 19 | 24 | 10 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 169 | 15 | 12 | 10 | 16 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 10 | 1 | |||||||||||
| 170 | 15 | 12 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 171 | 15 | 12 | 10 | 17 | 24 | 10 | 11 | 12 | 11 | 11 | 9 | 12 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 172 | 15 | 12 | 10 | 19 | 24 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 173 | 15 | 12 | 11 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 174 | 15 | 12 | 11 | 16 | 25 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 175 | 15 | 12 | 11 | 17 | 24 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 176 | 15 | 12 | 11 | 17 | 25 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 177 | 15 | 12 | 11 | 18 | 24 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 18 | 1 | ||||||||||
| 178 | 15 | 12 | 11 | 18 | 24 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 179 | 15 | 12 | 11 | 18 | 24 | 11 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 2 | |||||||||||
| 180 | 15 | 12 | 11 | 18 | 24 | 9 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 181 | 15 | 12 | 11 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 182 | 15 | 12 | 11 | 18 | 26 | 11 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 183 | 15 | 12 | 11 | 19 | 23 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 184 | 15 | 12 | 11 | 19 | 24 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 185 | 15 | 12 | 9 | 16 | 24 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 186 | 15 | 12 | 9 | 17 | 24 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 187 | 15 | 12 | 9 | 18 | 24 | 10 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 188 | 16 | 12 | 10 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 2 | |||||||||||
| 189 | 16 | 12 | 10 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 1 | 1 | ||||||||||
| 190 | 16 | 12 | 11 | 18 | 25 | 11 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 191 | 15 | 12 | 10 | 17 | 23 | 10 | 11 | 13 | 12 | 12 | 9 | 10 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 192 | 15 | 12 | 10 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 12 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 193 | 16 | 12 | 10 | 16 | 24 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 9 | 12 | 1 | |||||||||||
| 194 | 16 | 12 | 10 | 16 | 24 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 195 | 16 | 12 | 10 | 16 | 24 | 11 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 196 | 16 | 12 | 10 | 16 | 25 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 3 | |||||||||||
| 197 | 16 | 12 | 10 | 16 | 25 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 2 | |||||||||||
| 198 | 16 | 12 | 10 | 16 | 25 | 11 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 3 | |||||||||||
| 199 | 16 | 12 | 10 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 4 | 2 | 1 | ||||||||
| 200 | 16 | 12 | 10 | 18 | 22 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 201 | 16 | 12 | 10 | 19 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | ||||||||||
| 202 | 16 | 12 | 11 | 16 | 25 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 2 | |||||||||||
| 203 | 16 | 12 | 11 | 16 | 26 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 204 | 16 | 12 | 11 | 17 | 23 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 205 | 17 | 12 | 10 | 15 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 3 | |||||||||||
| 206 | 17 | 12 | 10 | 16 | 25 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 207 | 17 | 12 | 11 | 17 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 208 | 14 | 12 | 10 | 16 | 23 | 10 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 209 | 16 | 12 | 10 | 16 | 23 | 10 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 210 | 16 | 12 | 10 | 16 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 211 | 16 | 13 | 10 | 16 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 212 | 17 | 12 | 10 | 16 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 3 | |||||||||||
| 213 | 17 | 12 | 10 | 17 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 214 | 15 | 12 | 11 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 12 | 12 | 8 | 10 | 9 | 1 | |||||||||||
| 215 | 17 | 12 | 10 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 216 | 15 | 12 | 9 | 17 | 26 | 11 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 217 | 16 | 12 | 10 | 17 | 23 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | ||||||||||
| 218 | 16 | 12 | 10 | 18 | 25 | 10 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 219 | 13 | 12 | 11 | 15 | 22 | 10 | 15 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 220 | 15 | 12 | 10 | 17 | 25 | 12 | 8 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 221 | 15 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 222 | 15 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 1 | |||||||||||
| 223 | 15 | 12 | 11 | 16 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 224 | 15 | 13 | 10 | 17 | 26 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 12 | 11 | 1 | |||||||||||
| 225 | 16 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 5 | ||||||||||
| 226 | 16 | 12 | 10 | 18 | 26 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | ||||||||||
| 227 | 16 | 12 | 10 | 18 | 26 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | 1 | ||||||||||
| 228 | 15 | 12 | 10 | 16 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 229 | 16 | 12 | 10 | 17 | 26 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 230 | 16 | 12 | 11 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 231 | 16 | 12 | 11 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 10 | 11 | 10 | 1 | |||||||||||
| 232 | 15 | 12 | 10 | 17 | 21 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 233 | 15 | 12 | 10 | 17 | 22 | 10 | 15 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 234 | 15 | 12 | 10 | 17 | 24 | 11 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 235 | 15 | 12 | 10 | 17 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 6 | |||||||||||
| 236 | 15 | 12 | 10 | 17 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 237 | 15 | 12 | 10 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 10 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 238 | 15 | 12 | 10 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 3 | |||||||||||
| 239 | 15 | 12 | 10 | 18 | 25 | 11 | 11 | 15 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 240 | 15 | 12 | 11 | 16 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 241 | 16 | 12 | 10 | 16 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 242 | 16 | 12 | 10 | 17 | 24 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 243 | 16 | 12 | 10 | 17 | 24 | 11 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 244 | 16 | 12 | 10 | 17 | 24 | 12 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 245 | 16 | 12 | 10 | 18 | 24 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 2 | |||||||||||
| 246 | 16 | 12 | 11 | 17 | 24 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 247 | 16 | 12 | 11 | 17 | 24 | 11 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 248 | 16 | 12 | 11 | 18 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 249 | 16 | 12 | 9 | 16 | 24 | 12 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 250 | 14 | 12 | 11 | 18 | 23 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 251 | 15 | 12 | 10 | 17 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 12 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 252 | 15 | 12 | 10 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 253 | 15 | 12 | 10 | 18 | 25 | 10 | 11 | 13 | 13 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 254 | 15 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 13 | 9 | 11 | 12 | 8 | 11 | 10 | 2 | |||||||||||
| 255 | 15 | 12 | 10 | 19 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 9 | 1 | |||||||||||
| 256 | 15 | 12 | 11 | 16 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 12 | 11 | 1 | |||||||||||
| 257 | 15 | 12 | 11 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 258 | 15 | 12 | 11 | 17 | 25 | 11 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 1 | |||||||||||
| 259 | 15 | 12 | 11 | 18 | 24 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 2 | |||||||||||
| 260 | 15 | 12 | 11 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 261 | 15 | 12 | 11 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 262 | 15 | 13 | 9 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 2 | |||||||||||
| 263 | 16 | 12 | 10 | 17 | 25 | 12 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 9 | 1 | |||||||||||
| 264 | 16 | 12 | 10 | 17 | 26 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 265 | 16 | 12 | 10 | 18 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 266 | 16 | 12 | 10 | 18 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 267 | 16 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 2 | |||||||||||
| 268 | 16 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 12 | 12 | 9 | 11 | 10 | 1 | |||||||||||
| 269 | 16 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 13 | 9 | 11 | 12 | 8 | 11 | 10 | 6 | |||||||||||
| 270 | 16 | 12 | 10 | 18 | 25 | 11 | 11 | 14 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 271 | 16 | 12 | 10 | 19 | 25 | 11 | 11 | 13 | 12 | 13 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 272 | 16 | 12 | 11 | 16 | 24 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 273 | 16 | 12 | 11 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 274 | 16 | 12 | 11 | 18 | 24 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 275 | 16 | 12 | 11 | 18 | 25 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 276 | 16 | 12 | 11 | 18 | 25 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 1 | |||||||||||
| 277 | 16 | 12 | 11 | 18 | 25 | 11 | 11 | 13 | 12 | 10 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 278 | 16 | 12 | 11 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 279 | 16 | 12 | 9 | 17 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 280 | 17 | 12 | 10 | 17 | 24 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 281 | 17 | 12 | 10 | 18 | 23 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 282 | 17 | 12 | 10 | 18 | 24 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 2 | |||||||||||
| 283 | 17 | 12 | 10 | 18 | 25 | 10 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 284 | 17 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 285 | 17 | 12 | 10 | 18 | 25 | 11 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 11 | 10 | 1 | |||||||||||
| 8: | ||||||||||||||||||||||||||||
| 286 | 17 | 12 | 10 | 17 | 21 | 10 | 11 | 14 | 10 | 11 | 10 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 287 | 15 | 12 | 10 | 18 | 21 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 12 | 12 | 1 | |||||||||||
| 288 | 15 | 12 | 11 | 17 | 21 | 10 | 11 | 14 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 289 | 15 | 12 | 10 | 17 | 21 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 290 | 15 | 12 | 10 | 18 | 21 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 291 | 15 | 12 | 10 | 17 | 21 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 292 | 15 | 12 | 10 | 18 | 21 | 9 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 12 | 12 | 1 | |||||||||||
| 293 | 17 | 12 | 9 | 17 | 21 | 10 | 11 | 14 | 10 | 11 | 10 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 9: | ||||||||||||||||||||||||||||
| 294 | 14 | 14 | 11 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | 1 | 2 | 1 | ||||||||
| 295 | 14 | 15 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 296 | 14 | 15 | 10 | 17 | 22 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | 1 | |||||||||||
| 297 | 14 | 15 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 12 | 1 | |||||||||||
| 298 | 14 | 15 | 11 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 11 | 11 | 1 | |||||||||||
| 299 | 14 | 16 | 10 | 16 | 23 | 10 | 10 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 300 | 14 | 16 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 301 | 14 | 15 | 10 | 16 | 25 | 11 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 302 | 15 | 16 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 12 | 1 | |||||||||||
| 303 | 13 | 15 | 9 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 13 | 8 | 9 | 12 | 1 | |||||||||||
| 304 | 14 | 13 | 10 | 17 | 23 | 10 | 10 | 14 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 13 | 1 | |||||||||||
| 305 | 14 | 14 | 10 | 16 | 22 | 10 | 11 | 12 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 2 | |||||||||||
| 306 | 14 | 14 | 10 | 16 | 22 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 2 | |||||||||||
| 307 | 14 | 15 | 10 | 16 | 22 | 10 | 11 | 12 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 308 | 14 | 15 | 10 | 16 | 22 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 2 | |||||||||||
| 309 | 14 | 15 | 10 | 16 | 22 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 310 | 14 | 15 | 10 | 16 | 22 | 10 | 11 | 12 | 9 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 311 | 14 | 15 | 10 | 16 | 22 | 10 | 11 | 12 | 9 | 11 | 9 | 12 | 12 | 8 | 11 | 13 | 1 | |||||||||||
| 312 | 14 | 15 | 10 | 16 | 22 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 313 | 14 | 15 | 10 | 17 | 22 | 10 | 11 | 10 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 314 | 14 | 15 | 11 | 17 | 22 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 2 | |||||||||||
| 315 | 14 | 15 | 11 | 18 | 22 | 9 | 11 | 12 | 12 | 11 | 10 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 316 | 14 | 15 | 9 | 17 | 22 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 317 | 14 | 16 | 10 | 16 | 24 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 318 | 14 | 16 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 319 | 14 | 16 | 10 | 17 | 23 | 12 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 320 | 14 | 16 | 10 | 17 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 321 | 14 | 16 | 10 | 17 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 322 | 14 | 16 | 11 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 2 | 1 | ||||||||||
| 323 | 14 | 16 | 9 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 324 | 14 | 17 | 10 | 18 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 10 | 13 | 8 | 10 | 11 | 1 | |||||||||||
| 325 | 15 | 12 | 10 | 16 | 25 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 326 | 15 | 15 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 327 | 15 | 16 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 328 | 14 | 14 | 10 | 16 | 24 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | 2 | |||||||||||
| 329 | 14 | 15 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 330 | 15 | 15 | 12 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 331 | 15 | 15 | 9 | 16 | 24 | 10 | 11 | 12 | 10 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 332 | 15 | 15 | 9 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 2 | |||||||||||
| 333 | 15 | 15 | 11 | 16 | 23 | 10 | 11 | 14 | 12 | 11 | 10 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 334 | 15 | 15 | 9 | 17 | 23 | 9 | 11 | 12 | 14 | 11 | 9 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 335 | 17 | 13 | 11 | 16 | 24 | 11 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 336 | 14 | 14 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 337 | 14 | 14 | 9 | 19 | 23 | 9 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 14 | 1 | |||||||||||
| 338 | 14 | 15 | 11 | 16 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | 3 | ||||||||||
| 339 | 15 | 14 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 340 | 15 | 15 | 10 | 15 | 24 | 10 | 11 | 14 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 10 | 2 | |||||||||||
| 341 | 14 | 12 | 12 | 17 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 342 | 14 | 13 | 9 | 16 | 23 | 10 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 343 | 14 | 14 | 10 | 16 | 25 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 344 | 14 | 15 | 10 | 18 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 345 | 14 | 15 | 11 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 346 | 14 | 15 | 9 | 16 | 24 | 11 | 12 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 347 | 14 | 14 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 348 | 14 | 15 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 10 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 349 | 14 | 15 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 350 | 14 | 15 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 10 | 11 | 12 | 9 | 9 | 12 | 2 | |||||||||||
| 351 | 14 | 15 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 352 | 14 | 15 | 10 | 19 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 353 | 14 | 15 | 11 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 10 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 354 | 14 | 15 | 11 | 17 | 23 | 9 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 355 | 14 | 15 | 11 | 17 | 26 | 10 | 10 | 13 | 12 | 11 | 9 | 12 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 356 | 14 | 15 | 11 | 18 | 25 | 10 | 11 | 13 | 12 | 11 | 8 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 357 | 14 | 15 | 11 | 18 | 26 | 10 | 11 | 13 | 12 | 11 | 8 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 358 | 14 | 15 | 11 | 18 | 26 | 10 | 11 | 13 | 12 | 11 | 8 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 359 | 14 | 15 | 11 | 18 | 26 | 10 | 11 | 14 | 12 | 11 | 8 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 360 | 14 | 16 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 10 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 361 | 14 | 18 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 12 | 12 | 9 | 9 | 11 | 2 | |||||||||||
| 362 | 15 | 15 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 10 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 363 | 15 | 15 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 12 | 3 | |||||||||||
| 364 | 15 | 15 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 13 | 2 | |||||||||||
| 365 | 15 | 15 | 10 | 16 | 23 | 10 | 12 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 13 | 1 | |||||||||||
| 366 | 15 | 15 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 13 | 1 | |||||||||||
| 367 | 15 | 15 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 12 | 1 | |||||||||||
| 368 | 15 | 15 | 9 | 14 | 25 | 10 | 11 | 12 | 12 | 11 | 8 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 369 | 15 | 15 | 9 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 9 | 13 | 1 | |||||||||||
| 370 | 15 | 16 | 10 | 17 | 23 | 11 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 10 | 1 | |||||||||||
| 371 | 15 | 16 | 9 | 14 | 25 | 10 | 11 | 12 | 12 | 11 | 8 | 11 | 12 | 9 | 9 | 11 | 4 | |||||||||||
| 372 | 16 | 15 | 10 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 12 | 2 | |||||||||||
| 373 | 14 | 14 | 10 | 17 | 24 | 10 | 11 | 12 | 13 | 11 | 7 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 374 | 14 | 15 | 10 | 18 | 23 | 10 | 11 | 10 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 375 | 14 | 15 | 11 | 18 | 24 | 9 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 376 | 15 | 15 | 11 | 18 | 23 | 10 | 11 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 377 | 15 | 16 | 9 | 16 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 378 | 16 | 12 | 10 | 17 | 23 | 11 | 11 | 13 | 11 | 11 | 9 | 11 | 11 | 10 | 10 | 12 | 1 | |||||||||||
| 379 | 13 | 15 | 10 | 15 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 380 | 14 | 13 | 10 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 12 | 11 | 8 | 10 | 11 | 1 | |||||||||||
| 381 | 14 | 13 | 9 | 15 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 12 | 11 | 8 | 10 | 11 | 1 | |||||||||||
| 382 | 14 | 14 | 11 | 16 | 23 | 10 | 11 | 12 | 10 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 383 | 14 | 14 | 11 | 16 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 384 | 14 | 14 | 11 | 17 | 24 | 10 | 11 | 12 | 10 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 385 | 14 | 15 | 10 | 16 | 24 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 386 | 14 | 15 | 10 | 17 | 22 | 9 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 387 | 14 | 16 | 10 | 18 | 23 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 388 | 14 | 17 | 10 | 19 | 23 | 10 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 389 | 15 | 15 | 10 | 16 | 23 | 11 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 390 | 15 | 15 | 10 | 17 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 391 | 15 | 15 | 11 | 17 | 23 | 11 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | 1 | |||||||||||
| 392 | 15 | 15 | 11 | 17 | 23 | 11 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 9 | 13 | 1 | |||||||||||
| 393 | 15 | 15 | 11 | 17 | 24 | 11 | 11 | 13 | 14 | 11 | 9 | 11 | 12 | 8 | 9 | 13 | 1 | |||||||||||
| 394 | 15 | 15 | 9 | 16 | 22 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 395 | 15 | 15 | 9 | 16 | 24 | 10 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 9 | 9 | 12 | 1 | |||||||||||
| 396 | 15 | 15 | 9 | 17 | 24 | 11 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 397 | 16 | 15 | 11 | 16 | 23 | 10 | 11 | 12 | 12 | 11 | 9 | 12 | 12 | 8 | 9 | 13 | 1 | |||||||||||
| 398 | 16 | 15 | 9 | 16 | 23 | 9 | 12 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 399 | 16 | 15 | 9 | 16 | 23 | 9 | 12 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 400 | 16 | 15 | 9 | 17 | 24 | 10 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | 1 | |||||||||||
| 10: | ||||||||||||||||||||||||||||
| 401 | 15 | 13 | 10 | 15 | 25 | 10 | 11 | 14 | 12 | 11 | 11 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 402 | 15 | 13 | 10 | 16 | 25 | 10 | 11 | 14 | 12 | 11 | 11 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 403 | 15 | 13 | 10 | 17 | 23 | 10 | 11 | 15 | 12 | 11 | 11 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 404 | 15 | 13 | 11 | 16 | 22 | 9 | 11 | 14 | 12 | 11 | 11 | 11 | 12 | 8 | 10 | 11 | 5 | |||||||||||
| 405 | 16 | 13 | 10 | 15 | 22 | 9 | 11 | 14 | 12 | 11 | 11 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 406 | 16 | 13 | 11 | 16 | 22 | 9 | 11 | 14 | 12 | 11 | 11 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 407 | 16 | 14 | 10 | 16 | 25 | 10 | 11 | 13 | 12 | 11 | 11 | 11 | 12 | 8 | 10 | 10 | 4 | |||||||||||
| 408 | 16 | 14 | 10 | 17 | 25 | 10 | 11 | 13 | 12 | 11 | 11 | 11 | 12 | 8 | 10 | 10 | 3 | |||||||||||
| 12: | ||||||||||||||||||||||||||||
| 409 | 15 | 12 | 11 | 16 | 25 | 10 | 15 | 14 | 12 | 11 | 8 | 12 | 12 | 8 | 10 | 13 | 1 | |||||||||||
| 13: | ||||||||||||||||||||||||||||
| 410 | 17 | 12 | 9 | 15 | 25 | 11 | 13 | 12 | 11 | 11 | 9 | 12 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 21: | ||||||||||||||||||||||||||||
| 411 | 13 | 12 | 11 | 17 | 23 | 9 | 11 | 14 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 412 | 14 | 12 | 10 | 17 | 24 | 10 | 10 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 413 | 14 | 12 | 10 | 17 | 24 | 10 | 12 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | 1 | |||||||||||
| 414 | 14 | 12 | 10 | 17 | 24 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 415 | 15 | 12 | 10 | 17 | 24 | 10 | 11 | 13 | 10 | 11 | 9 | 11 | 13 | 8 | 10 | 12 | 1 | |||||||||||
| 416 | 13 | 12 | 9 | 19 | 24 | 10 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 417 | 17 | 12 | 9 | 17 | 21 | 10 | 11 | 14 | 10 | 11 | 10 | 11 | 12 | 8 | 11 | 11 | 1 | |||||||||||
| 418 | 13 | 12 | 10 | 19 | 24 | 10 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 419 | 13 | 12 | 10 | 19 | 24 | 11 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 420 | 13 | 12 | 11 | 17 | 24 | 11 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 421 | 13 | 12 | 9 | 19 | 24 | 10 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 9 | 9 | 13 | 1 | |||||||||||
| 422 | 15 | 12 | 11 | 17 | 24 | 9 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | 1 | |||||||||||
| 423 | 13 | 12 | 10 | 17 | 24 | 10 | 11 | 13 | 10 | 11 | 9 | 11 | 11 | 8 | 10 | 12 | 1 | |||||||||||
| 424 | 13 | 12 | 10 | 17 | 24 | 10 | 11 | 13 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 425 | 13 | 12 | 10 | 18 | 22 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 426 | 13 | 12 | 9 | 17 | 22 | 10 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | 1 | |||||||||||
| 427 | 13 | 12 | 9 | 17 | 22 | 10 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 26: | ||||||||||||||||||||||||||||
| 428 | 15 | 12 | 9 | 18 | 21 | 10 | 11 | 15 | 15 | 10 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 429 | 14 | 13 | 9 | 17 | 23 | 10 | 10 | 14 | 12 | 11 | 9 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 430 | 16 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 431 | 16 | 12 | 9 | 17 | 23 | 10 | 12 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 10 | 13 | 3 | |||||||||||
| 432 | 14 | 10 | 9 | 16 | 23 | 10 | 14 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 433 | 14 | 12 | 10 | 17 | 23 | 10 | 12 | 13 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | 1 | |||||||||||
| 434 | 15 | 12 | 9 | 16 | 22 | 10 | 14 | 12 | 11 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 1 | |||||||||||
| 435 | 15 | 12 | 9 | 16 | 25 | 10 | 13 | 14 | 11 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | 2 | |||||||||||
| 436 | 15 | 12 | 9 | 17 | 25 | 10 | 13 | 14 | 11 | 11 | 9 | 11 | 12 | 8 | 11 | 10 | 1 | |||||||||||
| 28: | ||||||||||||||||||||||||||||
| 437 | 14 | 12 | 10 | 14 | 22 | 10 | 12 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 13 | 1 | |||||||||||
| 438 | 14 | 12 | 9 | 15 | 22 | 10 | 13 | 11 | 13 | 11 | 10 | 11 | 12 | 9 | 10 | 11 | 4 | |||||||||||
| 439 | 14 | 12 | 9 | 15 | 22 | 10 | 15 | 11 | 13 | 11 | 10 | 11 | 12 | 9 | 10 | 11 | 1 | |||||||||||
| 440 | 14 | 12 | 9 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 10 | 11 | 12 | 8 | 11 | 12 | 1 | |||||||||||
| 441 | 14 | 12 | 9 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 10 | 11 | 13 | 1 | |||||||||||
| 442 | 14 | 12 | 9 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 13 | 4 | 1 | 2 | |||||||||
| 443 | 14 | 12 | 9 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 444 | 14 | 12 | 9 | 16 | 22 | 11 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 14 | 1 | |||||||||||
| 445 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | 1 | ||||||||||
| 446 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 447 | 15 | 12 | 10 | 16 | 23 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 448 | 15 | 12 | 10 | 16 | 22 | 11 | 15 | 13 | 13 | 11 | 9 | 11 | 13 | 10 | 10 | 13 | 1 | |||||||||||
| 449 | 15 | 12 | 10 | 17 | 22 | 10 | 15 | 13 | 13 | 11 | 9 | 11 | 13 | 10 | 10 | 13 | 1 | |||||||||||
| 450 | 13 | 12 | 9 | 16 | 25 | 11 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 13 | 1 | |||||||||||
| 451 | 14 | 12 | 11 | 18 | 22 | 10 | 14 | 11 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 13 | 1 | |||||||||||
| 452 | 14 | 12 | 9 | 15 | 22 | 10 | 14 | 11 | 13 | 11 | 10 | 11 | 12 | 9 | 10 | 11 | 1 | 1 | ||||||||||
| 453 | 14 | 12 | 9 | 16 | 22 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 13 | 1 | |||||||||||
| 454 | 14 | 12 | 9 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | 1 | |||||||||||
| 455 | 14 | 12 | 9 | 16 | 23 | 10 | 10 | 15 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 13 | 1 | |||||||||||
| 456 | 15 | 12 | 10 | 16 | 23 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 9 | 13 | 1 | |||||||||||
| 457 | 15 | 12 | 10 | 17 | 23 | 10 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 458 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 459 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 12 | 12 | 10 | 10 | 13 | 3 | |||||||||||
| 460 | 15 | 13 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 2 | |||||||||||
| 461 | 16 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 4 | |||||||||||
| 462 | 16 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 11 | 12 | 1 | |||||||||||
| 463 | 16 | 12 | 11 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 464 | 16 | 12 | 11 | 16 | 23 | 10 | 15 | 13 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 465 | 17 | 12 | 11 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 466 | 17 | 12 | 11 | 16 | 22 | 11 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 9 | 12 | 1 | |||||||||||
| 467 | 14 | 12 | 11 | 16 | 22 | 10 | 15 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 7 | |||||||||||
| 468 | 14 | 12 | 11 | 16 | 23 | 10 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 469 | 14 | 12 | 12 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 470 | 15 | 12 | 11 | 16 | 23 | 10 | 11 | 12 | 13 | 11 | 9 | 11 | 13 | 10 | 10 | 12 | 1 | |||||||||||
| 471 | 16 | 13 | 11 | 16 | 24 | 11 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 472 | 14 | 12 | 10 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 11 | 13 | 1 | |||||||||||
| 473 | 15 | 12 | 10 | 17 | 24 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 474 | 15 | 12 | 11 | 16 | 24 | 10 | 15 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 475 | 15 | 12 | 9 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 11 | 13 | 1 | |||||||||||
| 476 | 15 | 13 | 11 | 15 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 477 | 14 | 12 | 10 | 15 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | 1 | |||||||||||
| 478 | 15 | 12 | 10 | 17 | 23 | 10 | 13 | 10 | 10 | 10 | 9 | 11 | 11 | 10 | 10 | 11 | 1 | |||||||||||
| 479 | 15 | 12 | 10 | 17 | 23 | 10 | 13 | 10 | 10 | 10 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 480 | 15 | 12 | 11 | 17 | 23 | 10 | 12 | 14 | 10 | 10 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 481 | 15 | 12 | 11 | 17 | 23 | 10 | 13 | 10 | 10 | 10 | 9 | 11 | 12 | 10 | 10 | 11 | 5 | |||||||||||
| 482 | 15 | 12 | 11 | 17 | 23 | 10 | 13 | 10 | 10 | 10 | 9 | 11 | 12 | 10 | 11 | 11 | 1 | |||||||||||
| 483 | 15 | 12 | 11 | 17 | 23 | 10 | 13 | 10 | 10 | 10 | 9 | 11 | 12 | 10 | 11 | 12 | 1 | |||||||||||
| 484 | 15 | 12 | 11 | 17 | 23 | 10 | 13 | 10 | 10 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | 2 | |||||||||||
| 485 | 15 | 12 | 11 | 17 | 23 | 11 | 13 | 10 | 10 | 10 | 9 | 11 | 12 | 10 | 10 | 11 | 2 | |||||||||||
| 486 | 15 | 12 | 11 | 17 | 29 | 10 | 13 | 10 | 10 | 10 | 9 | 11 | 12 | 10 | 10 | 11 | 1 | |||||||||||
| 487 | 13 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 488 | 14 | 12 | 9 | 15 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 489 | 14 | 12 | 9 | 16 | 23 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 490 | 14 | 12 | 9 | 17 | 21 | 10 | 16 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | 1 | |||||||||||
| 491 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 10 | 12 | 12 | 10 | 10 | 12 | 1 | |||||||||||
| 492 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 10 | 12 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 493 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 10 | 11 | 12 | 2 | |||||||||||
| 494 | 15 | 12 | 11 | 16 | 22 | 10 | 14 | 12 | 12 | 12 | 9 | 11 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 495 | 15 | 12 | 9 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 496 | 16 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 10 | 11 | 12 | 10 | 10 | 13 | 1 | |||||||||||
| 497 | 16 | 13 | 9 | 16 | 22 | 11 | 13 | 12 | 12 | 11 | 9 | 11 | 12 | 9 | 11 | 14 | 1 | |||||||||||
| 498 | 14 | 12 | 9 | 16 | 22 | 10 | 14 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 12 | 1 | |||||||||||
| 499 | 14 | 12 | 9 | 17 | 22 | 10 | 11 | 13 | 13 | 11 | 9 | 11 | 12 | 9 | 11 | 12 | 1 | |||||||||||
| 500 | 15 | 12 | 10 | 15 | 22 | 10 | 14 | 13 | 13 | 11 | 9 | 11 | 13 | 10 | 10 | 13 | 1 | |||||||||||
| 501 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 12 | 12 | 10 | 11 | 12 | 1 | |||||||||||
| 502 | 15 | 12 | 10 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 12 | 12 | 9 | 10 | 13 | 1 | |||||||||||
| 503 | 16 | 12 | 9 | 16 | 22 | 10 | 14 | 12 | 13 | 11 | 9 | 12 | 12 | 10 | 10 | 12 | 1 | |||||||||||
Population codes are as defined in table 1.
Table 5.
Y-STR Haplotypes, Found in Five or More Individuals, and Their Haplogroup Designation
| DYS19; DYS388; DYS389I; DYS389b; DYS390; DYS391;DYS392; DYS393; DYS425; DYS426; DYS434; DYS437;DYS435; DYS438; DYS436; DYS439 | Haplogroup | Population (No. of Individuals)a |
| 14_12_10_17_23_11_13_14_12_12_9_9_11_10_12_12 | 1 | HZR (6) |
| 14_12_11_16_23_10_10_14_12_12_9_10_11_11_12_12 | 1 | PRS (13) |
| 16_12_9_16_23_10_11_14_14_11_9_10_11_10_12_11 | 2 | KAL (6) |
| 15_12_10_17_24_11_11_13_12_12_9_8_11_11_12_10 | 3 | PKH (6) |
| 15_12_11_18_24_10_11_12_12_12_9_8_11_11_12_11 | 3 | BRU (18) MAKB (1) |
| 16_12_9_17_25_11_11_14_12_12_9_8_11_11_12_11 | 3 | BAL (4); MAKB (1); MAKN (1); SDH (3) |
| 16_12_10_17_24_11_11_13_12_12_9_8_11_11_12_10 | 3 | BAL (1); BSK (1); PKH (10); SDH (1) |
| 16_12_10_17_25_11_11_13_12_12_9_8_11_11_12_10 | 3 | BSK (1); KSR (4); PKH (2); SDH (1) |
| 16_12_10_18_25_10_11_13_12_12_9_8_11_11_12_10 | 3 | BLT (2); KSR (1); PRS (2); SDH (1) |
| 16_12_10_18_25_11_11_13_12_12_9_8_11_11_12_10 | 3 | MAKN (1); SDH (5) |
| 16_12_10_18_25_11_11_13_12_13_9_8_11_11_12_10 | 3 | SDH (6) |
| 14_14_11_16_23_10_11_12_12_11_9_8_11_9_12_12 | 9 | BAL (1); BRU (1); KAL (2); MAKB (1) |
| 15_13_11_16_22_9_11_14_12_11_11_8_11_10_12_11 | 10 | BSK (5) |
| 16_12_10_16_22_10_14_12_13_11_9_10_11_10_12_12 | 26/28 | BSK (5) |
| 14_12_9_16_22_10_14_11_13_11_9_9_11_10_12_13 | 28 | BAL (4); MAKB (1); SDH (2) |
| 14_12_11_16_22_10_15_12_13_11_9_10_11_10_12_12 | 28 | KAL (7) |
| 15_12_11_17_23_10_13_10_10_10_9_10_11_10_12_11 | 28 | PRS (5) |
Population codes are as defined in figure 1.
The GST and modal size of the repeat unit, for all 16 Y-STRs examined in the Pakistani population, are given in table 6. The correlation between marker heterozygosity and GST was found not to be significant (r=0.329; P=.213). The modal size and variance of the 16 Y-STRs within haplogroups 1, 2, 3, 8, 9, 10, 21, 26, and 28 is also given in table 6. Certain haplogroups have a different modal allele size, and some examples of this are shown in boldface italics in table 6. For instance, DYS388 has 15 repeats in haplogroup 9, compared with 12 repeats in most of the other haplogroups in Pakistan. Similarly, the modal allele for DYS438 is 9 in haplogroup 9, but 10 or 11 in the other haplogroups. The modal allele for DYS434 for haplogroup 10 is 11, which is strikingly different from the allele size of this locus in other haplogroups. The complete lack of variability for DYS436 in the 233 male subjects belonging to haplogroup 3 is notable. Haplogroup 10 appears to have the least variability across most loci except for DYS390 (table 6). These findings demonstrate the strong structuring of Y-STR variability by haplogroup.
Table 6.
Modal Allele ± Variance and GST of the 16 Y-STRs within Each Haplogroup and in the Pakistani Population
|
Modal Allele ± Variance in |
|||||||||||
| Haplogroup |
|||||||||||
| Y-STR | GST | 1a | 2b | 3c | 8d | 9e | 10f | 21g | 26h | 28i | Pakistanj |
| DYS19 | .073 | 14 ± .4 | 15 ± .8 | 16 ± .5 | 15 ± .9 | 14 ± .4 | 16 ± .3 | 13 ± 1.2 | 15 ± .6 | 15 ± .6 | 15 ± .9 |
| DYS388 | .056 | 12 ± .2 | 12 ± .2 | 12 ± .0 | 12 ± .0 | 15 ± .9 | 13 ± .3 | 12 ± .0 | 12 ± .4 | 12 ± .0 | 12 ± 1.5 |
| DYS389I | .089 | 11 ± .4 | 10 ± .7 | 10 ± .3 | 10 ± .3 | 10 ± .5 | 10 ± .2 | 10 ± .5 | 9 ± .2 | 10 ± .7 | 10 ± .5 |
| DYS389b | .117 | 16 ± .7 | 16 ± 1.3 | 18 ± .7 | 17 ± .3 | 16 ± .8 | 16 ± .4 | 17 ± .8 | 17 ± .4 | 16 ± .4 | 16 ± 1.0 |
| DYS390 | .085 | 23 ± .7 | 23 ± .5 | 25 ± .6 | 21 ± .0 | 23 ± .8 | 25 ± 2.2 | 24 ± 1.0 | 23 ± 1.6 | 22 ± .8 | 23 ± 1.4 |
| DYS391 | .085 | 10 ± .3 | 10 ± .4 | 11 ± .3 | 10 ± .4 | 10 ± .2 | 10 ± .3 | 10 ± .3 | 10 ± .0 | 10 ± .1 | 10 ± .3 |
| DYS392 | .090 | 10 ± 3.5 | 11 ± .5 | 11 ± .3 | 11 ± .0 | 11 ± .1 | 11 ± .0 | 11 ± .2 | 12 ± 1.5 | 14 ± 1.1 | 11 ± 1.8 |
| DYS393 | . 131 | 14 ± .5 | 12 ± .5 | 13 ± .3 | 13 ± .3 | 12 ± .4 | 14 ± .4 | 13 ± .2 | 12 ± 1.3 | 12 ± .9 | 13 ± .8 |
| DYS425 | .095 | 12 ± .3 | 12 ± 1.0 | 12 ± .0 | 12 ± .7 | 12 ± .5 | 12 ± .0 | 10 ± 1.2 | 12 ± 1.3 | 13 ± 1.2 | 12 ± .6 |
| DYS426 | .088 | 12 ± .2 | 11 ± .1 | 12 ± .1 | 11 ± .0 | 11 ± .1 | 11 ± .0 | 11 ± .1 | 11 ± .1 | 11 ± .2 | 11 ± .3 |
| DYS434 | .124 | 9 ± .0 | 9 ± .2 | 9 ± .0 | 9 ± .2 | 9 ± .2 | 11 ± .0 | 9 ± .1 | 9 ± .0 | 9 ± .1 | 9 ± .2 |
| DYS437 | .126 | 10 ± .7 | 8 ± 1.0 | 8 ± .1 | 8 ± .0 | 9 ± .5 | 8 ± .0 | 8 ± .1 | 9 ± .6 | 10 ± .3 | 8 ± .7 |
| DYS435 | .028 | 11 ± .1 | 11 ± .1 | 11 ± .0 | 11 ± .0 | 11 ± .1 | 11 ± .0 | 11 ± .0 | 11 ± .0 | 11 ± .1 | 11 ± .1 |
| DYS438 | .119 | 11 ± .3 | 10 ± .3 | 11 ± .3 | 11 ± .2 | 9 ± .3 | 10 ± .2 | 10 ± .1 | 10 ± .3 | 10 ± .1 | 11 ± .7 |
| DYS436 | .074 | 12 ± .1 | 12 ± .2 | 12 ± .0 | 12 ± .0 | 12 ± .0 | 12 ± .0 | 12 ± .1 | 12 ± .0 | 12 ± .1 | 12 ± .1 |
| DYS439 | .067 | 12 ± 1.0 | 11 ± .5 | 10 ± .3 | 12 ± .7 | 12 ± .6 | 11 ± .5 | 12 ± .3 | 12 ± .7 | 12 ± .7 | 11 ± 1.0 |
Total number of Y-STR haplotypes found = 88; number of chromosomes found = 130.
Total number of Y-STR haplotypes found = 60; number of chromosomes found = 70.
Total number of Y-STR haplotypes found = 137; number of chromosomes found = 233.
Total number of Y-STR haplotypes found = 8; number of chromosomes found = 8.
Total number of Y-STR haplotypes found = 107; number of chromosomes found = 132.
Total number of Y-STR haplotypes found = 8; number of chromosomes found = 17.
Total number of Y-STR haplotypes found = 17; number of chromosomes found = 17.
Total number of Y-STR haplotypes found = 9; number of chromosomes found = 12.
Total number of Y-STR haplotypes found = 67; number of chromosomes found = 97.
Total number of Y-STR haplotypes found = 503; number of chromosomes found = 718 (includes one individual from each of haplogroups 12 and 13).
We wanted to calculate a Y-based measure of genetic distance between populations that would reflect the differentiation that had occurred within Pakistan and that would not be disproportionately dominated by ancient differences that had previously accumulated between haplogroups. The standard way to do this would be to use STR variation, and table 7 summarizes population pairwise values of ΦST on the basis of STR variation alone (A) or of binary-marker plus STR variation (B), with binary-marker differences weighted 10 times higher than STR differences. These matrices are highly correlated (r=0.95; P<.001), as might be expected from the structuring of STR variation by haplogroup. However, these measures are significantly influenced by ancient differences, and we have therefore developed a modified measure. We reasoned that much of the STR variation within haplogroups would have originated recently and could be used for this purpose. We therefore calculated population pairwise values of ΦST, on the basis of STR variation within haplogroups, and used a weighted average of these to produce a single ΦST distance matrix (table 7C; fig. 3). These distances are also highly correlated with distances based on STRs alone (r=0.76; P<.001) or on STRs plus binary markers (r=0.70; P<.001), but a greater proportion of the variation is seen between populations (22%, compared with 6% and 7%, respectively). A comparison of figure 3 with figure 2B (which was based on binary marker frequencies alone) reveals a striking overall resemblance, with the Hazaras being distinct from all of the other populations. The other outstanding populations are the Kalash and Parsis (as before), the Kashmiris (perhaps because of the small sample), and the Brahuis, who are thus more distinct in their STR profiles than haplogroup frequencies. MDS plots of the distances in tables 7A and 7B (not shown) lead to similar conclusions, but resemble figure 2B more closely in the way that the Brahuis do not stand out so much.
Table 7.
Population Pairwise ΦST Values Based on STR Variation Alone, Combined Binary Marker and STR Variation, or Weighted Within-Haplogroup STR Variation
|
Population Pairwise ΦST Values Based on STR Variation Alone |
|||||||||||
| Population | Baluch | Balti | Brahui | Burusho | Hazara | Kalash | Kashmiri | Makrani Baluch | Makrani Negroid | Parsi | Pathan |
| Balti | .027 | ||||||||||
| Brahui | .053 | .055 | |||||||||
| Burusho | .036 | .012 | .084 | ||||||||
| Hazara | .092 | .124 | .158 | .126 | |||||||
| Kalash | .063 | .104 | .120 | .049 | .195 | ||||||
| Kashmiri | .084 | .006 | .083 | .114 | .142 | .216 | |||||
| Makrani Baluch | .004 | .007 | .021 | .032 | .080 | .078 | .053 | ||||
| Makrani Negroid | .013 | .001 | .043 | .033 | .091 | .088 | .044 | −.011 | |||
| Parsi | .078 | .092 | .076 | .091 | .142 | .107 | .154 | .052 | .074 | ||
| Pathan | .049 | .001 | .078 | .054 | .113 | .126 | .026 | .032 | .034 | .135 | |
| Sindhi | .065 | −.010 | .054 | .069 | .129 | .14 | −.002 | .027 | .021 | .122 | .024 |
|
Population Pairwise ΦST Values Based on Combined Binary Marker and STR Variation |
|||||||||||
| Baluch |
Balti |
Brahui |
Burusho |
Hazara |
Kalash |
Kashmiri |
Makrani Baluch |
Makrani Negroid |
Parsi |
Pathan |
|
| Balti | .005 | ||||||||||
| Brahui | .048 | .017 | |||||||||
| Burusho | .021 | −.010 | .058 | ||||||||
| Hazara | .107 | .134 | .129 | .095 | |||||||
| Kalash | .052 | .095 | .078 | .060 | .159 | ||||||
| Kashmiri | .088 | −.020 | .072 | .087 | .201 | .234 | |||||
| Makrani Baluch | −.005 | −.020 | .003 | .015 | .083 | .052 | .056 | ||||
| Makrani Negroid | .014 | .000 | .013 | .031 | .086 | .055 | .065 | −.020 | |||
| Parsi | .078 | .101 | .061 | .100 | .132 | .066 | .198 | .037 | .053 | ||
| Pathan | .033 | −.020 | .050 | .028 | .133 | .108 | .025 | .021 | .031 | .140 | |
| Sindhi | .057 | −.020 | .028 | .050 | .141 | .126 | −.001 | .017 | .023 | .125 | .014 |
|
Population Pairwise ΦST Values Based on Weighted Within-Haplogroup STR Variation |
|||||||||||
| Baluch |
Balti |
Brahui |
Burusho |
Hazara |
Kalash |
Kashmiri |
Makrani Baluch |
Makrani Negroid |
Parsi |
Pathan |
|
| Balti | .164 | ||||||||||
| Brahui | .174 | .311 | |||||||||
| Burusho | .212 | .127 | .348 | ||||||||
| Hazara | .454 | .653 | .545 | .520 | |||||||
| Kalash | .337 | .305 | .351 | .233 | .642 | ||||||
| Kashmiri | .173 | .143 | .426 | .271 | .483 | .455 | |||||
| Makrani Baluch | .004 | .094 | .151 | .208 | .463 | .344 | .173 | ||||
| Makrani Negroid | .035 | .091 | .239 | .167 | .516 | .271 | .156 | .008 | |||
| Parsi | .285 | .173 | .260 | .283 | .558 | .402 | .330 | .169 | .175 | ||
| Pathan | .174 | .261 | .361 | .203 | .490 | .248 | .226 | .211 | .158 | .346 | |
| Sindhi | .059 | .039 | .268 | .129 | .503 | .195 | .073 | .054 | .017 | .117 | .140 |
Figure 3.
Multidimensional scaling presentation of weighted population pairwise values of ΦST. RSQ value = 0.81. Linguistic affiliations of populations are indicated as in figure 2B.
Median-Joining Networks
The genetic relationships among the different Pakistani ethnic groups were explored further by drawing median-joining networks (Bandelt et al. 1995), and examples are shown in figures 4, 5, and 6. The haplogroup 1 network (fig. 4) reveals considerable variation, but also a high degree of population-specific substructure. For example, the 24 Parsi haplogroup 1 chromosomes all fall into one of three clusters (fig. 4, green), 19 of 26 Burusho haplogroup 1 chromosomes fall into two clusters (blue), and 12 of 14 Hazara haplogroup 1 chromosomes fall into a single cluster, and all of these clusters are specific to their respective populations. The haplogroup 10 network (fig. 5) is much simpler, because of the smaller number of chromosomes, but again reveals population-specific clustering for Burusho and Hazara haplotypes. The haplogroup 28 network (fig. 6) shows a striking isolated Parsi-specific cluster, at the end of a long branch, containing 15 of 16 Parsi haplogroup 28 chromosomes. Clusters of Kalash, Burusho, and—to a lesser degree—Baluch chromosomes are also evident, although one Baluch haplotype is shared with Sindhi and Makrani Baluch individuals from nearby southern populations.
Figure 4.
Median-joining network of haplogroup 1 individuals, based on their Y-STR haplotypes. Circles represent haplotypes and have an area proportional to frequency, except the circle designated 013+, which represents 13 Parsi individuals and has been reduced in size. Color represents the population of origin. STR differences are shown on the lines linking haplotypes.
Figure 5.
Median-joining network of haplogroup 10 individuals, based on their Y-STR haplotypes. Conventions used are as in figure 4.
Figure 6.
Median-joining network of haplogroup 28 individuals, based on their Y-STR haplotypes. Conventions used are as in figure 4, except that some STR differences are shown by colored lines to make a complex region of the network clearer.
BATWING TMRCAs were calculated for the haplogroup 28 network and for selected lineages within a number of haplogroups. The results are summarized in table 8.
Table 8.
BATWING Estimates of TMRCAs for Selected Lineages
| Haplogroup | Lineage within Haplogroup | PopulationSubdivision | Mode TMRCA [95% CI](years) |
| 28 | Entire haplogroup | Yes | 7,200 [4,400–14,000] |
| 28 | Entire haplogroup | No | 6,900 [4,000–13,000] |
| 28 | Parsi-specific | No | 1,800 [600–4,500] |
| 1 | Hazara-specific | No | 400 [120–1,200] |
| 10 | Hazara-specific | No | 100 [6–600] |
Discussion
We have carried out the first extensive analysis of Y diversity within Pakistan, examining 34 markers in 718 male subjects from 12 populations. This allows us to compare Pakistani Y diversity with that previously reported in world populations, to investigate differences within Pakistan, and to evaluate some of the suggested population histories from a Y perspective.
Comparisons with Worldwide Data
In a worldwide comparison, Pakistani populations mostly cluster around a pooled South Asian sample and lie close to a Middle Eastern sample (fig. 2A). This finding is unsurprising, in part because the South Asian sample included 62 Pakistani individuals (i.e., 32% of 196 total) and in part because Y variation in many areas of the world is predominantly structured by geography, not by language or ethnic affiliation (Rosser et al. 2000; Zerjal et al. 2001). The greater genetic similarity of Pakistani populations to those in the west than to eastern populations is illustrated by the fact that four of the five frequent haplogroups in Pakistan (haplogroups 1, 2, 3, and 9, which together make up 79% of the total population) are also frequent in western Asia and Europe but not in China or Japan; conversely, the haplogroups that are frequent in East Asia (e.g., 4, 5, 10, 13, and 20) are rare or absent in Pakistan, forming only 2.5% of the total. If, as in some interpretations, an early exodus from Africa along the southern coast of Asia led to the first anatomically modern human populations in Pakistan, and these people carried the eastern haplogroups or their precursors, their Y chromosomes have now been largely replaced by subsequent migrations or gene flow; indeed, the representatives of the eastern haplogroups in Pakistan may be derived from modern back-migration, not from ancient survivors.
The fifth haplogroup that is common in Pakistan, haplogroup 28, differs from all the others in its distribution. Within Pakistan, it made up 14% of our sample and was present in all but two populations (both of which had very small sample sizes), so it is both common and widespread. Outside Pakistan and the nearby countries, however, it is rare. It has been reported in India (30%; present in 3/3 populations), Tajikistan (10%; present in 5/6 populations), and Uzbekistan (3%; present in 10/13 populations), but it is rare in Russia (0.4%; present in 1/6 populations) and the Caucasus (1.4%; present in 1/6 populations (Wells et al. 2001) and has not been found at all in China or Mongolia (unpublished observations). BATWING estimates of the TMRCA of the Pakistani haplogroup 28 chromosomes were ∼7,000 (4,000–14,000) years (table 8). Thus, within this time period, the Pakistani populations have diverged from a common ancestral population or have experienced considerable male gene flow between themselves or from a common source. Since the estimated age corresponds to the early Neolithic period, the spread of this lineage might be associated with the local expansion of farmers.
Comparisons within Pakistan
Haplogroup distributions in Pakistani populations, with the exception of the Hazara (discussed in the next section), are strikingly similar to one another (figs. 1 and 2), despite some notable linguistic differences. Indeed, the language isolate-speaking Burusho, the Dravidian-speaking Brahuis, and the Sino-Tibetan–speaking Baltis did not stand out from the other populations at all in the haplogroup analyses (table 2 and fig. 2), suggesting either that the linguistic differences arose after the common Y pattern was established or that there has been sufficient Y gene flow between populations to eliminate any initial differences. Yet a more detailed analysis of the Y haplotypes (e.g., figs. 3–6) reveals some distinct features of the Brahui and considerable population specificity; population-specific clusters of related haplotypes are commonly found in these networks. Such clusters will only be seen if populations are isolated from one another. It may be that a low degree of gene flow between populations over a long time is sufficient to result in similar haplogroup frequencies without producing many shared clusters.
Population-specific clusters of haplotypes are particularly evident in some populations. In the Hazaras, where the distinct haplogroup frequencies noted above are found, most chromosomes (19/23; 83%) fall into one of just two well-isolated clusters (figs. 4 and 5), whereas the Parsis, the Kalash, and the Burusho also show prominent clusters. The Hazaras, Parsis, and Kalash were the three populations showing the most significantly different population pairwise ΦST values. The high values of the Hazaras and Parsis can partly be accounted for by migration to Pakistan from other places, but a contributing factor is likely to be drift, either due to a limited number of founder lineages or occurring subsequently within small populations. Θk values (Ewens 1972) provide a way of comparing effective population sizes. Values based on the STRs for the Hazaras, Parsis, Kalash, and Burushos were 8.9, 77.5, 25.8, and 74.2, respectively, compared with a mean of 181.8 for the other populations with sample sizes >20. Effective population size for Y chromosomes can differ greatly from census population size, but it is notable that the Parsis and Kalash do have the smallest census sizes, one-hundredth or one-thousandth of most of those of the other populations (table 1), so these small census sizes may have been maintained for a long time. In summary, many features of the present Pakistani Y haplotype distributions can be accounted for by a shared ancestral gene pool, with limited gene flow between populations and drift in the smaller ones.
Insights into Population Origins
The suggested population origins (table 1) can now be considered in the light of these Y results. Information is provided by haplogroup frequencies, which can be used to produce admixture estimates, and these are easy to interpret if populations are large and isolated and the source populations have different frequencies. When these conditions are not met, the presence of distinct Y lineages can still be informative. The origins of the Parsis are well-documented (Nanavutty 1997) and thus provide a useful test case. They are followers of the Iranian prophet Zoroaster, who migrated to India after the collapse of the Sassanian empire in the 7th century a.d. They settled in 900 a.d. in Gujarat, India, where they were called the “Parsi” (meaning “from Iran”). Eventually they moved to Mumbai in India and Karachi in Pakistan, from where the present population was sampled (fig. 7). Their frequencies for haplogroups 3 (8%) and 9 (39%) do indeed resemble those in Iran more than those of their current neighbors in Pakistan. They show the lowest frequency for haplogroup 3 in Pakistan (apart from the Hazaras; fig. 1C). The mean for eight Iranian populations was 14% (n=401) (Quintana-Murci et al. 2001), whereas that for Pakistan, excluding the Parsis, was 36%. The corresponding figures for haplogroup 9 were 39% in the Parsis, 40% in Iran, and 15% in Pakistan excluding the Parsis. These figures lead to an admixture estimate of 100% from Iran (table 3). Given the small effective population size of the Parsis, the closeness of their match to the Iranian data may be fortuitous, and the presence of haplogroup 28 chromosomes at 18% (4% in Iran; Wells et al. 2001) suggests some gene flow from the surrounding populations. The TMRCA for the Parsi-specific cluster in the haplogroup 28 networks was 1,800 (600–4,500) years (table 8), consistent with the migration of a small number of lineages from Iran. Overall, these results demonstrate a close match between the historical records and the Y data, and thus suggest that the Y data will be useful when less historical information is available.
Figure 7.
Traditions of population origin supported by Y data. Solid arrows, movements also supported by historical data. Dashed arrows, movements also supported by oral traditions. Arrows indicate the country of origin or continent of origin (Negroid Makrani) but not the precise geographical location or route. Darker boundaries represent disputed borders.
The population that is genetically most distinct, the Hazaras, claims descent from Genghis Khan’s army; their name is derived from the Persian word “hazar,” meaning “thousand,” because troops were left behind in detachments of a thousand. Toward the end of the 19th century, some Hazaras moved from Afghanistan to the Khurram Valley in Pakistan, the source of the samples investigated here. Thus, their oral history identifies an origin in Mongolia and population bottlenecks ∼800 and ∼100 years ago. Of the two predominant Y haplogroups present in this population, haplogroup 1 is widespread in Pakistan, much of Asia, Europe, and the Americas, and so provides little information about the place of origin. Haplogroup 10, in contrast, is rare in most Pakistani populations (1.4%, when the Hazaras are excluded) but is common in East Asia, including Mongolia, where it makes up over half of the population (unpublished results). Admixture estimates (table 3) are consistent with a substantial contribution from Mongolia. BATWING analysis of the Hazara-specific haplotype clusters in haplogroups 1 and 10 suggested TMRCAs of 400 (120–1,200) and 100 (6–600) years (table 8), respectively. Thus, the genetic evidence is consistent with the oral tradition and, in view of its independent nature, provides strong support for it (fig. 7).
Some other suggested origins receive more limited support from the Y data. The Negroid Makrani, with a postulated origin in Africa, carry the highest frequency of haplogroup 8 chromosomes found in any Pakistani population, as noted elsewhere (Qamar et al. 1999). This haplogroup is largely confined to sub-Saharan Africa, where it constitutes about half of the population (Hammer et al. 2001) and can thus be regarded as a marker of African Y chromosomes. Nevertheless, it makes up only 9% of the Negroid Makrani sample, and haplogroup 28 (along with other typical Pakistani haplogroups) is present in this population. If the Y chromosomes were initially African (fig. 7), most have subsequently been replaced: the overall estimate of the African contribution is ∼12% (table 3).
The Balti are thought to have originated in Tibet, where the predominant haplogroups are 4 and 26. Neither was present in the sample from this study, providing no support for a Tibetan origin of the Y chromosome lineages and an admixture estimate of zero (table 3). However, this result must be interpreted with caution, because of the small sample size. Three populations have possible origins from the armies of Alexander the Great: the Burusho, the Kalash, and the Pathans. Modern Greeks show a moderately high frequency of haplogroup 21 (28%; Rosser et al. 2000), but this haplogroup was not seen in either the Burusho or the Kalash sample and was found in only 2% of the Pathans, whereas the local haplogroup 28 was present at 17%, 25%, and 13%, respectively. Greek-admixture estimates of 0% were obtained for the Burusho and the Pathans, but figures of 20%–40% were observed for the Kalash (table 3). In view of the absence of haplogroup 21, we ascribe this result either to drift in the frequencies of the other haplogroups, particularly haplogroups 2 and 1, or to the poor resolution of lineages within these haplogroups, resulting in distinct lineages being classified into the same paraphyletic haplogroups. Overall, no support for a Greek origin of their Y chromosomes was found, but this conclusion does require the assumption that modern Greeks are representative of Alexander’s armies. Two populations, the Kashmiris and the Pathans, also lay claim to a possible Jewish origin. Jewish populations commonly have a moderate frequency of haplogroup 21 (e.g., 20%) and a high frequency of haplogroup 9 (e.g., 36%; (Hammer et al. 2000). The frequencies of both of these haplogroups are low in the Kashmiris and Pathans, and haplogroup 28 is present at 13% in the Pathans, so no support for a Jewish origin is found, and the admixture estimate was 0% (table 3), although, again, this conclusion is limited both by the small sample size available from Kashmir and by the assumption that the modern samples are representative of ancient populations.
The suggested origin of the Baluch is in Syria. Syrians, like Iranians, are characterized by a low frequency of haplogroup 3 and a high frequency of haplogroup 9 (9% and 57%, respectively; Hammer et al. 2000), whereas the corresponding frequencies in the Baluch are 29% and 12%. This difference and the high frequency of haplogroup 28 in the Baluch (29%) make a predominantly Syrian origin for their Y chromosome unlikely, and the admixture estimate was 0% (table 3), although the 8% frequency for haplogroup 21, the highest identified in Pakistan thus far, does indicate some western contribution to their Y lineages. The Brahuis have a possible origin in West Asia (Hughes-Buller 1991) and it has been suggested that a spread of haplogroup 9 Y chromosomes was associated with the expansion of Dravidian-speaking farmers (Quintana-Murci et al. 2001). Brahuis have the highest frequency of haplogroup 9 chromosomes in Pakistan (28%) after the Parsis, providing some support for this hypothesis, but their higher frequency of haplogroup 3 (39%) is not typical of the Fertile Crescent (Quintana-Murci et al. 2001) and suggests a more complex origin, possibly with admixture from later migrations, such as those of Indo-Iranian speakers from the steppes of Central Asia and others from further east. This possibility is supported by the detection of low frequencies of haplogroups 10, 12, and 13 in the Brahuis, all rare in Pakistan and typical of East Asia, East and northern Asia, and Southeast Asia, respectively.
The failure to find a Y link with a suggested population of origin does not disprove a historical association, but it does demonstrate that the Y chromosomes derived from such historical events have been lost or replaced. Analyses of mitochondrial DNA and other loci would help to elucidate the population histories and would be particularly interesting in populations like the Negroid Makrani and the Balti, in which there is a contrast between the phenotype and the typical Pakistani Y haplotypes.
Acknowledgments
This work was supported by a Wellcome Trust Collaborative Research Initiative Grant to S.Q.M. T.Z. was also supported by The Wellcome Trust, and C.T.-S. by the Cancer Research Campaign. We express our appreciation to the original DNA donors who made this study possible. The Department of Health of the Government of Baluchistan and the Baluch Student Federation, Quetta, Pakistan, assisted in the collection of the Brahui and Baluch samples. Pathan samples were collected with the assistance of the Department of Paediatrics, Lady Reading Post Graduate Medical Hospital, Peshawar, Pakistan. We are also grateful to Dr. I. Kazmi and the Aga Khan Foundation Rural Health Support Program for their assistance in the collection of Burusho samples. Dr. F. Sethna provided valuable assistance in the collection of the Parsi samples. We thank Luis Quintana-Murci for his comments on the manuscript.
Electronic-Database Information
URLs for data in this article are as follows:
- Arlequin, http://anthropologie.unige.ch/arlequin/
- BATWING, http://www.maths.abdn.ac.uk/~ijw/
- Network 2.0, http://www.fluxus-engineering.com/
- ViSta, http://forrest.psych.unc.edu/
References
- Ahmad AKN (1952) Jesus in heaven on earth. The Civil and Military Gazette Ltd, Lahore, Pakistan [Google Scholar]
- Ayub Q, Mohyuddin A, Qamar R, Mazhar K, Zerjal T, Mehdi SQ, Tyler-Smith C (2000) Identification and characterisation of novel human Y-chromosomal microsatellites from sequence database information. Nucleic Acids Res 28:e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom PC (1992) Balti. In: Backstrom PC, Radloff CF (eds) Sociolinguistic survey of northern Pakistan. Vol 2, Languages of northern areas. National Institute of Pakistan Studies, Islamabad, pp 3–27 [Google Scholar]
- Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellew HW (1979) The races of Afghanistan. Sang-e-Meel Publications, Lahore, Pakistan [Google Scholar]
- Bellew HW (1998) An enquiry into the ethnography of Afghanistan. Vanguard Books, Lahore, Pakistan [Google Scholar]
- Bergen AW, Wang C-Y, Tsai J, Jefferson K, Dey C, Smith KD, Park S-C, Tsai S-J, Goldman D (1999) An Asian–Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet 63:63–80 [DOI] [PubMed] [Google Scholar]
- Bianchi NO, Bailliet G, Bravi CM, Carnese RF, Rothhammer F, Martinez-Marignac VL, Pena SD (1997) Origin of Amerindian Y-chromosomes as inferred by the analysis of six polymorphic markers. Am J Phys Anthropol 102:79–89 [DOI] [PubMed] [Google Scholar]
- Biddulph J (1977) Tribes of the Hindoo Koosh. Indus Publications, Karachi, Pakistan [Google Scholar]
- Burton RF (1851) Sindh and the races that inhabit the valley of the Indus. WH Allen and Co Ltd, London [Google Scholar]
- Caroe O (1958) The Pathans. Oxford University Press, Karachi, Pakistan [Google Scholar]
- Casanova M, Leroy P, Boucekkine C, Weissenbach J, Bishop C, Fellous M, Purrello M, Fiori G, Siniscalco M (1985) A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 230:1403–1406 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Dales GF (1991) The phenomenon of the Indus civilization. In: Jansen M, Mulloy M, Urban G (eds) Forgotten cities on the Indus: early civilization in Pakistan from the 8th to the 2nd millennia BC. Verlag Philipp von Zabern, Mainz, Germany, pp 129–144 [Google Scholar]
- Decker KD (1992) Sociolinguistic survey of Northern Pakistan. Vol 5, Languages of Chitral. National Institute of Pakistan Studies, Islamabad [Google Scholar]
- Ewens WJ (1972) The sampling theory of selectively neutral alleles. Theor Popul Biol 3:87–112 [DOI] [PubMed] [Google Scholar]
- Grimes BF (1992) Ethnologue: languages of the world. Summer Institute of Linguistics, Dallas [Google Scholar]
- Hammer MF (1994) A recent insertion of an Alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol 11:749–761 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Karafet TM, Redd AJ, Jarjanazi H, Santachiara-Benerecetti S, Soodyall H, Zegura SL (2001) Hierarchical patterns of global human Y-chromosome diversity. Mol Biol Evol 18:1189–1203 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B (2000) Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurdardottir S, Nicholson J, Sykes B, Hill EW, Bradley DG, Bosnes V, Gulcher JR, Ward R, Stefansson K (2000) Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 67:697–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes-Buller R (1991) Imperial gazetteer of India: provincial series, Baluchistan. Sang-e-Meel, Lahore, Pakistan [Google Scholar]
- Hussain J (1997) A history of the peoples of Pakistan towards independence. Oxford University Press, Karachi, Pakistan [Google Scholar]
- Ibbetson D (1883) Panjab castes. Sang-e-Meel, Lahore, Pakistan [Google Scholar]
- Jarrige JF (1991) Mehrgarh: its place in the development of ancient cultures in Pakistan. In: Jansen M, Mulloy M, Urban G (eds) Forgotten cities on the Indus: early civilization in Pakistan from the 8th to the 2nd millennia BC. Verlag Philipp von Zabern, Mainz, Germany, pp 34–50 [Google Scholar]
- Jobling MA, Tyler-Smith C (2000) New uses for new haplotypes: the human Y chromosome, disease and selection. Trends Genet 16:356–362 [DOI] [PubMed] [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Kruger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Tyler-Smith C, Mendonca BB, Hughes I, Berkovitz GD, Goodfellow PN, Hawkins JR (1996) Mutation analysis of the 2 kb 5′ to SRY in XY females and XY intersex subjects. J Med Genet 33:465–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC (1991) The genetic structure of admixed populations. Genetics 127:417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias N, Bayes M, Tyler-Smith C (1994) Highly informative compound haplotypes for the human Y chromosome. Hum Mol Genet 3:115–123 [DOI] [PubMed] [Google Scholar]
- Mehdi SQ, Qamar R, Ayub Q, Kaliq S, Mansoor A, Ismail M, Hammer MF, Underhill PA, Cavalli-Sforza LL (1999) The origins of Pakistani populations: evidence from Y chromosome markers. In: Papiha SS, Deka R, Chakraborty R (eds) Genomic diversity: applications in human population genetics. Kluwer Academic/Plenum Publishers, New York, pp 83–90 [Google Scholar]
- Mohyuddin A, Ayub Q, Qamar R, Zerjal T, Helgason A, Mehdi SQ, Tyler-Smith C (2001) Y-chromosomal STR haplotypes in Pakistani populations. Forensic Sci Int 118:141–146 [DOI] [PubMed] [Google Scholar]
- Nanavutty P (1997) The Parsis. National Book Trust, New Delhi, India [Google Scholar]
- Pandya A, King TE, Santos FR, Taylor PG, Thangaraj K, Singh L, Jobling MA, Tyler-Smith C (1998) A polymorphic human Y-chromosomal G to A transition found in India. Ind J Hum Genet 4:52–61 [Google Scholar]
- Qamar R, Ayub Q, Khaliq S, Mansoor A, Karafet T, Mehdi SQ, Hammer MF (1999) African and Levantine origins of Pakistani YAP+ Y chromosomes. Hum Biol 71:745–755 [PubMed] [Google Scholar]
- Quddus SA (1990) The tribal Baluchistan. Ferozsons (Pvt) Ltd, Lahore, Pakistan [Google Scholar]
- Quintana-Murci L, Krausz C, Zerjal T, Sayar SH, Hammer MF, Mehdi SQ, Ayub Q, Qamar R, Mohyuddin A, Radhakrishna U, Jobling MA, Tyler-Smith C, McElreavey K (2001) Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am J Hum Genet 68:537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DF, Hiorns R (1965) Methods of analysis of the genetic composition of a hybrid population. Hum Biol 37:38–43 [PubMed] [Google Scholar]
- Robertson GS (1896) The Kafirs of the Hindu-Kush. Oxford University Press, Karachi, Pakistan [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FR, Carvalho-Silva DR, Pena SDJ (1999a) PCR-based DNA profiling of human Y chromosomes. In: Epplen JT, Lubjuhn T (eds) Methods and tools in biosciences and medicine. Birkhauser Verlag, Basel, Switzerland, pp 133–152 [Google Scholar]
- Santos FR, Pandya A, Kayser M, Mitchell RJ, Liu A, Singh L, Destro-Bisol G, Novelletto A, Qamar R, Mehdi SQ, Adhikari R, Knijff P, Tyler-Smith C (2000) A polymorphic L1 retroposon insertion in the centromere of the human Y chromosome. Hum Mol Genet 9:421–430 [DOI] [PubMed] [Google Scholar]
- Santos FR, Pandya A, Tyler-Smith C, Pena SD, Schanfield M, Leonard WR, Osipova L, Crawford MH, Mitchell RJ (1999b) The central Siberian origin for native American Y chromosomes. Am J Hum Genet 64:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Kueffer J-M, Roessli D, Excoffier L (1997) Arlequin ver 1.1: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland [Google Scholar]
- Seielstad MT, Hebert JM, Lin AA, Underhill PA, Ibrahim M, Vollrath D, Cavalli-Sforza LL (1994) Construction of human Y-chromosomal haplotypes using a new polymorphic A to G transition. Hum Mol Genet 3:2159–1261 [DOI] [PubMed] [Google Scholar]
- Shinka T, Tomita K, Toda T, Kotliarova SE, Lee J, Kuroki Y, Jin DK, Tokunaga K, Nakamura H, Nakahori Y (1999) Genetic variations on the Y chromosome in the Japanese population and implications for modern human Y chromosome lineage. J Hum Genet 44:240–245 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Bradman N, Flinn HM (1999) High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet 105:577–581 [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C (1999) Y-chromosomal DNA markers. In: Papiha SS, Deka R, Chakraborty R (eds) Genomic diversity: applications in human population genetics. Kluwer Academic/Plenum Publishers, New York, pp 65–73 [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield LS, Sulston JE, Goodfellow PN (1995) Sequence variation of the human Y chromosome. Nature 378:379–380 [DOI] [PubMed] [Google Scholar]
- Wilson IJ, Balding DJ (1998) Genealogical inference from microsatellite data. Genetics 150:499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert S (2000) A new history of India. Oxford University Press, New York [Google Scholar]
- Young FW, Bann CM (1996) A visual statistics system. In: Stine RA, Fox J (eds) Statistical computing environments for social researches. Sage Publications, New York, pp 207–236 [Google Scholar]
- Zerjal T, Beckman L, Beckman G, Mikelsaar AV, Krumina A, Kucinskas V, Hurles ME, Tyler-Smith C (2001) Geographical, linguistic, and cultural influences on genetic diversity: Y-chromosomal distribution in Northern European populations. Mol Biol Evol 18:1077–1087 [DOI] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183 [PMC free article] [PubMed] [Google Scholar]