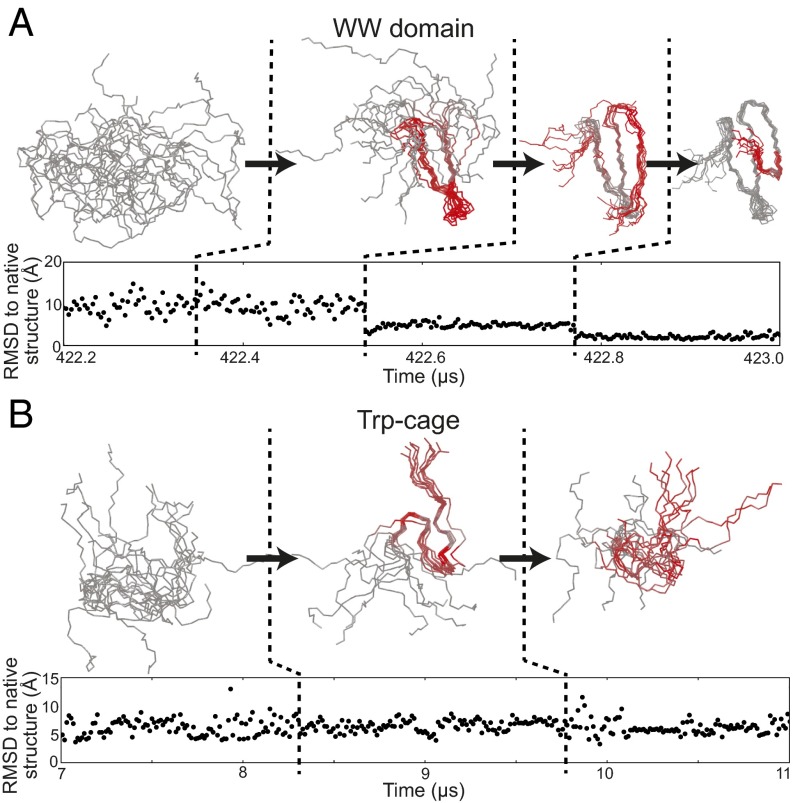
Fig. 2.
Results from application of SIMPLE to equilibrium protein-folding simulation trajectories. (A) Three consecutive detected conformational changes in the WW domain reveal a folding pathway. (B) Two consecutive detected conformational changes in Trp-cage during the unfolded state reveal transient formation of a metastable structure that does not lie along the overall folding pathway. Several of these changes are not evident in the time series of RMSD to the native structure, which is often used in the analysis of protein simulations. Aligned ensemble images are shown for the protein backbone structures between each successive pair of detected changes, with red color used to indicate the spatial location of the change between the current ensemble and the previous ensemble as indicated by SIMPLE’s output (SI Text). As inputs to SIMPLE, we used 595 interatomic distance observables across ∼65,000 time points for the WW domain trajectory and 190 interatomic distance observables across ∼21,000 time points for the Trp-cage trajectory (SI Text). Changes detected over the full lengths of these simulation trajectories are shown in Figs. S1 and S2. The average number of observables found to change distribution at each change point was 205 for the WW domain trajectory and 69 for the Trp-cage trajectory.