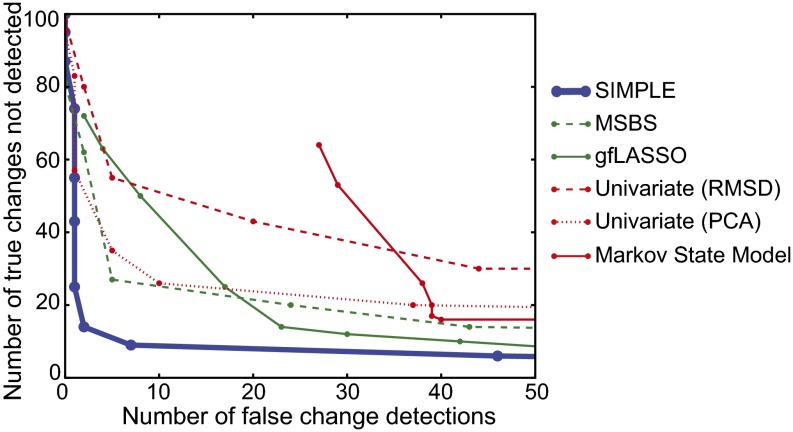
Fig. 4.
Comparison of the performance of various methods for detecting conformational changes in synthetic protein trajectories in which the conformational changes take place at predetermined times. SIMPLE (blue) outperforms several methods previously published in the biomolecular simulation literature (red), including a state identification method involving Markov state models (32) and a univariate change-point detection method (17) applied to time series of either the RMSD from native structure (“RMSD”) or the coefficient of the first principal component (“PCA”). SIMPLE also outperforms certain related change-point detection methods described in the recent statistics literature (green), including the group-fused LASSO (gfLASSO) approach of Bleakley and Vert (24) and an adaptation of the multisample binary segmentation (MSBS) approach of Zhang et al. (20).