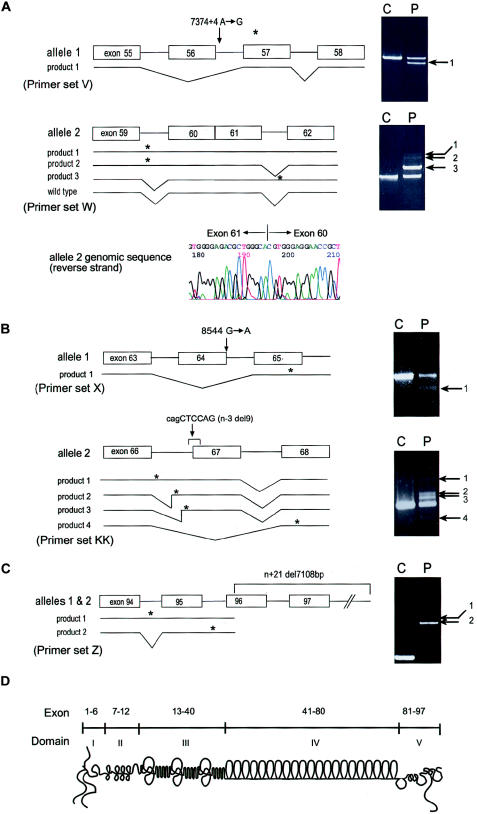
Figure 1.
Mutation analysis and schematic diagram of perlecan. A, Results of RT-PCR, which was performed with RNA from patient 1 fibroblasts, using primer sets V and W and analyzed on 1% agarose gels as described elsewhere (Arikawa-Hirasawa et al. 2001). Sequencing revealed that product 1 from allele 1 was missing the entire exon 56 sequence and that products 1–3 from allele 2 retained intron sequences. Product 3 of allele 2 contains the sequence of intron 61, whereas products 1 and 2 contain either both introns 59 and 61 or intron 59, respectively. Exon 56 skipping in allele 1 causes frameshift and is predicted to produce a premature termination codon in exon 57. The intron 59 retention of products 1 and 2 from allele 2 is predicted to produce a premature termination codon within intron 59, and the intron 61 retention of product 3 is predicted to produce a termination codon within intron 61. Genomic sequencing identified heterozygous mutations in an A→G transition at the +4 position (donor site) of intron 56 (7374+4 A→G) in allele 1. Allele 2 showed a complete loss of intron 60, resulting in fusion of exons 60 and 61 in the perlecan gene. The exon-fusion mutation created aberrant transcripts, products 1–3, but is also predicted to produce the wild-type product. Since primer set W does not distinguish the wild-type product derived from alleles 1 or 2, we used a primer set from exons 56 and 62 to exclude a wild-type transcript from allele 1, in which exon 56 is missing and confirmed the presence of the wild-type transcript together with aberrant splicing transcripts produced by allele 2 (data not shown). B, Patient 2. RT-PCR was performed with RNA from skeletal muscle tissues of patient 2, using primer sets X and KK. Sequencing of product 1 with primers X revealed exon 64 skipping, and products 1, 2, and 3 with primers KK contained total or partial retention of intron 66. Product 4 with primers KK showed exon 67 skipping. Genomic sequencing revealed a heterozygous mutation in a G→A transition at the donor site of exon 64 (8564 G→A) in allele 1 and a 9-bp deletion (cagCTCCAG) at the acceptor junction of intron 66 and exon 67 in allele 2 (n−3 del9). C, Patient 3. RT-PCR was performed with RNA from patient 3 fibroblasts using primer set Z. Product 1 contained sequences including introns 94 and 95. Product 2, a major product, contained intron 95. Sequencing of the genomic PCR product revealed a 7,108-bp deletion beginning at the 5′ portion of exon 96 and extending to the 3′ flanking sequence of HSPG2. The deletion results in aberrant splicing, including exon skipping and intron retention. Asterisk (*) indicates a premature termination codon. Arrows indicate abnormally sized RT-PCR products. D,Schematic diagram of perlecan and its exon and domain structure. Primer sequences for RT-PCR are as follows: V (forward, ATCACGGTCACAGTAACTGGGACC; reverse, CCTGCACCGTTACTGACGTG); KK (forward, ATGGCACAAGCGTGGAGGAAACC; reverse, AGGCTTCTTGCTCAGGGCCTGG); W (forward, ATCCAGCAGCGCCTTAGTGG; reverse, CATGCCCATCAGAATTGAGTCAT); X (forward, CTCAACAACATCG ATGCCCTGGAG; reverse, CTCCAGCCCAGGACCCATTCCT); and Z (forward, AGGCAAGGACTTCATCAGCCTCGGG; reverse, TCGACTTGGATGGAACCTCTGCGG).