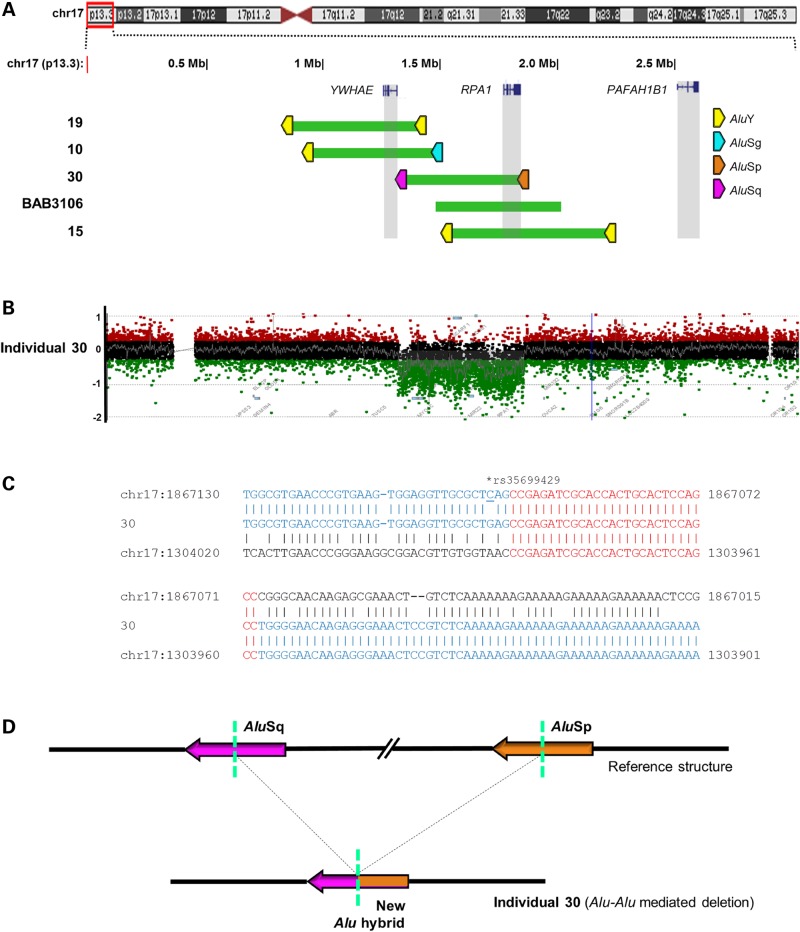
Figure 1.
Interstitial deletions of 17p13.3. (A) Deletions were plotted by exact genomic coordinates according to breakpoint junction sequencing results using the UCSC genome browser custom track function (GRCh37/hg19 assembly). Green blocks represent deletions. Upper panel shows an ideogram of human chromosome 17 with the 17p13.3 region involved highlighted in the red box. Three critical genes in 17p13.3 deletion and duplication syndromes, YWHAE, RPA1 and PAFAH1B1, are highlighted in gray vertical boxes. Alu pairs that mediated the deletions are denoted by left-facing (− strand) pentagons. Whether an Alu element presents on the plus or minus strand is defined according to the UCSC genome browser (GRCh37/hg19 assembly). Pentagons filled with different colors indicate different Alu families. (B, C) Representative aCGH log2 plot and breakpoint junction sequence of individual 30 with an interstitial deletion. Breakpoint junction sequence is aligned to the proximal and distal genomic references and color-matched. Microhomology at the breakpoint is indicated in red. (D) Proposed rearrangement of Alu–Alu-mediated deletion in individual 30. Array plots and breakpoint sequences of remaining interstitial deletions are shown in Supplementary Material, Figure S1.