Figure 3.
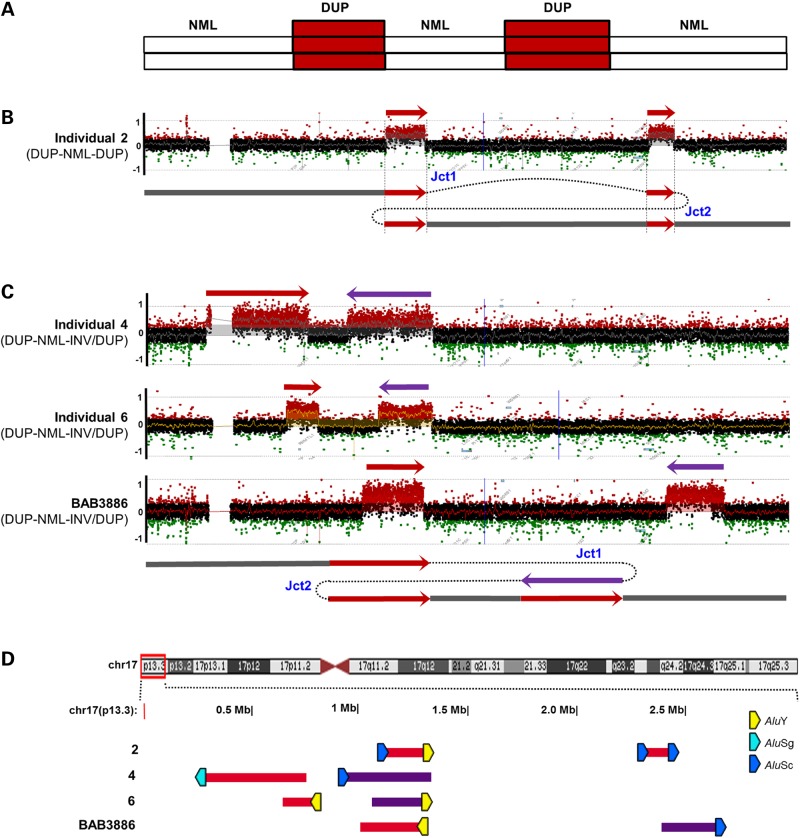
Complex rearrangements distinguished by a DUP–NML–DUP pattern of 17p13.3. (A) Illustration of the DUP–NML–DUP rearrangement pattern from aCGH results. (B) Array CGH log2 plot and proposed generation mechanism of DUP–NML–DUP rearrangement in individual 2. (C) Array CGH log2 plot and proposed generation mechanism of DUP–NML–INV/DUP rearrangement in individuals 4, 6 and BAB3886. (D) CNVs were plotted by exact genomic coordinates according to breakpoint junction sequencing results using the UCSC genome browser custom track function. Red blocks, duplication; Purple blocks, inverted duplication. Upper panel shows an ideogram of human chromosome 17 with the 17p13.3 region involved highlighted in the red box. Alu pairs that mediated the complex rearrangements are denoted by right-facing (+ strand) or left-facing (− strand) pentagons. Pentagons filled with different colors indicate different Alu families. Breakpoint sequences of all the DUP–NML–DUP individuals are shown in Supplementary Material, Figure S3. Jct1 and Jct2, junction 1 and junction 2.