Figure 3.
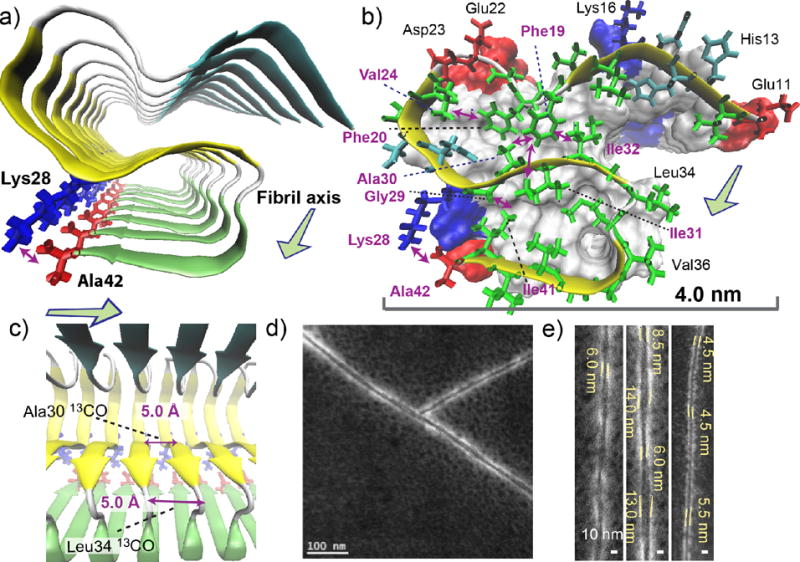
Structural details of the Aβ(1–42) fibril revealed by the SSNMR analysis. (a–c) A structural model of the amyloid fibril of Aβ(1–42). Disordered residues 1–10 were omitted for clarity. (a) View from the fibril axis shows three β-strand regions (arrows) connected by short coil (white) or turn (silver) regions (tube); the β-strands are represented by color-coded arrows in cyan (resides 12–18), yellow (24–33), and green (36–40). The unique salt bridge between Lys28 (blue) and Ala42 (red) is shown. (b) Side chain contacts for a single Aβ chain in a skeletal and a ribbon diagram with a van der Waals surface and polarity diagram for the rest of the Aβ chains. Hydrophobic, polar, acidic, and basic residues are represented by green, cyan, red, and blue, respectively. Observed long-range side-chain intra-molecular contacts (purple arrows) and inter-molecular contacts (blue arrow) are shown. All β-sheet regions are presented in yellow. The surface plot indicates positively charged (Lys; blue) and negatively charged (Glu, Asp; red) side chains, and Ala42 that has a negatively charged carboxyl group (red). (c) The side view in ribbon diagram. The in-register parallel β-sheet arrangement was confirmed by measurements of intermolecular 13CO-13CO distances of ∼4.8 Å at Ala30 and Leu34 (purple arrows). (d, e) Scanning TEM (STEM) images of seeded fibril filaments. (e) The diameters of the fibril filaments are ranged between 4.5 and 6.0 nm for thinner filaments (left and right) and between 6.0 and 14.0 nm for wider filaments (middle).
