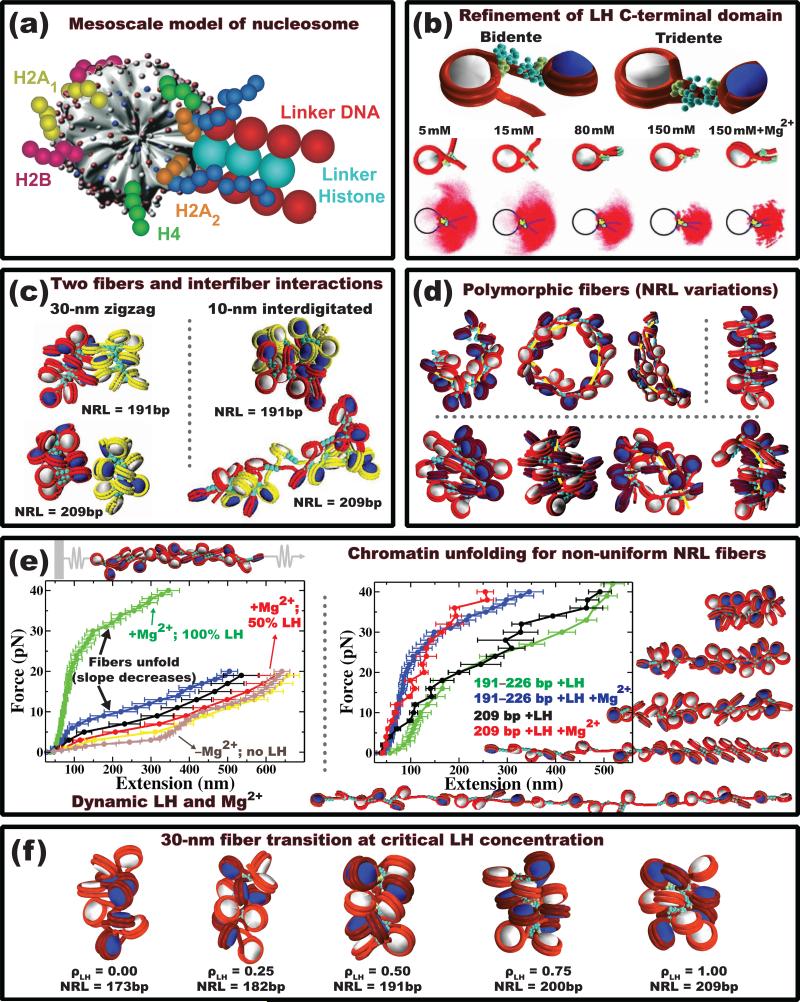
Figure 3.
Recent findings on chromatin fibers using variations of the Schlick group's mesoscale model. (a) The mesoscale model of nucleosome is composed of an irregularly shaped core with uniformly distributed 300 Debye-Hückel charges, 10 flexible core histone tails representing the C-termini of H2A and the N-termini of H2A/H2B/H3/H4, and linker DNA beads described as a worm-like chain; it can also include a linker histone attached to the dyad axis of the nucleosome [68,5•]. (b) A refined model captures the spontaneous condensation of the linker histone C-terminal domain upon nucleosome binding [69••]. The side view trajectories of the nucleosome exiting/entering linker DNA show increasing formation of the nucleosome stem as a function of the monovalent salt concentration and the presence of divalent salts in the environment. At low salt, the nucleosome adopts a bidente configuration (top left), where the linker histone interacts with the nucleosome core and the entry linker DNA, leading to the formation of extended fibers. At high salt, the nucleosome adopts a tridente configuration (top right), where the linker histone interacts with the nucleosome core and both linker DNAs, promoting the stem formation and overall fiber compaction. (c) Two chromatin fibers (red and yellow) can interact in various ways. Low chromatin densities favor the formation of segregated canonical zigzag 30-nm fibers with weak inter-fiber interactions. Denser chromatin environments lead to heteromorphic and metastable 10-nm interdigitated fibers stabilized by stronger inter-fiber interactions [11]. (d) Nonuniform (alternating) DNA linker lengths (measured in units of nucleosome repeat lengths, NRL) increase the structural diversity of chromatin fibers [12••]. Fibers with one short linker DNA length (26 bp) can form bent-ladder like structures (top left); fibers with medium to long linker DNAs (35 to 79 bp) and moderate NRL variations, NRL (see full details in Ref. [12••]), mostly adopt canonical zigzag conformations (top right); long linker length (>44 bp) fibers with large NRL variations are highly polymorphic as to adopt various stable equilibrium conformations ranging from canonical to slightly bent to intra-looped nature. (e) Various chromatin unfolding simulations help interpret force/extension curves measured by single-molecule manipulation experiments. These have helped interpret the effect of linker histones, divalent Mg2+ ions, and non-uniform nucleosome repeat lengths (NRL) on chromatin unfolding. Here, a strong negative correlation between forced extension and linker histone concentration is captured (left) [73]. Fibers with nonuniform NRLs exhibit a smoother structural transition than uniform NRL fibers (right) [75]. (f) The empirical relationship between linker histone concentration and NRL was recently associated with formation of compact 30-nm fibers. The critical linker histone concentration, ρLH (ratio of linker histones per nucleosome), for this transition increases linearly with the NRL [74].