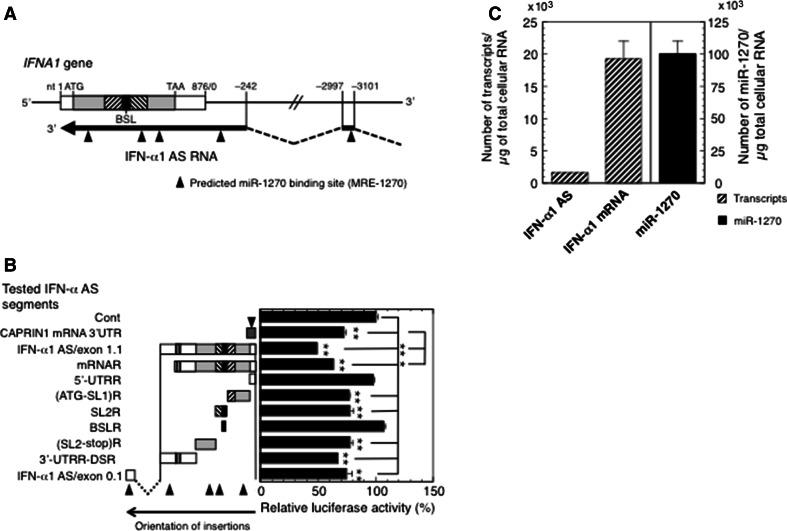
Fig. 3.
Characterization of MRE-1270s on IFN-α1 AS. a Location of predicted miR-1270 binding sites on IFN-α1 AS/exons 0.1 and 1.1. Bioinformatic analysis of miR-1270 binding sequences on IFN-α1 AS/exons 0.1 and 1.1 revealed the presence of five highly conserved predicted MRE-1270s (closed triangles). b Confirmation of IFN-α1 AS segments harboring the predicted MRE-1270s. A series of pEF-Luc reporters, into which the following IFN-α1 AS segments were inserted; IFN-α1 AS/exon 1.1, mRNA-revertant (mRNAR), 5′-UTRR, (ATG-SL1)R, SL2R, BSLR, (SL2-stop)R, 3′-UTRR-DSR, or IFN-α1 AS/exon 0.1 were transfected into pLKO-miR-1270-transduced Namalwa cells. pEF-Luc-CAPRIN1 3′-UTR, which harbors one MRE-1270, was included as a positive control for the assay. The luciferase activities were normalized and are presented as described in the legend to Fig. 1. The transfection efficiencies of each reporter were normalized to Renilla luciferase activity, an internal transfection standard. *p < 0.05, **p < 0.01. Closed triangles are as described in the legend to a. c Quantification of copy numbers of IFN-α1 AS/mRNA and miR-1270. Namalwa cells were infected with SeV and harvested for RT-qPCR analysis at 24 h after infection. Copy numbers of IFN-α1 AS/mRNA and miR-1270 were determined from the regression lines from the sets of standards as described in the “Materials and methods” section. The results are presented as the “Number of transcripts or miR-1270/µg of total cellular RNA” ±SEM of four or five samples. High levels of sequence homology between IFNA1 and IFNA13 (99.8 % in the coding region) did not allow distinction between these genes [11]. The expression levels of IFN-α1 mRNA and AS RNA presented in this study thus represent the total amounts of both IFN-α1 and -α13 mRNA/AS RNA