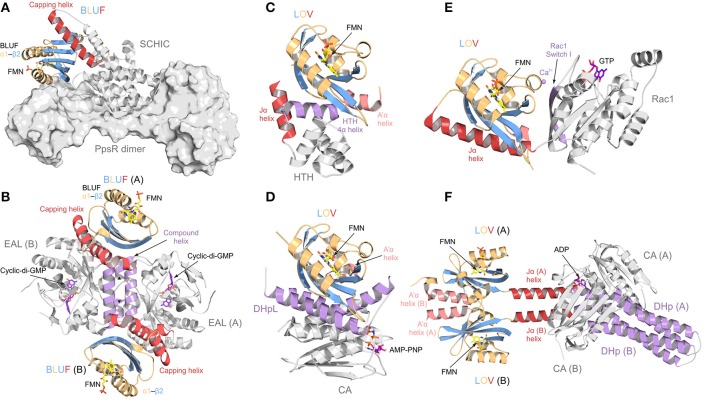
Figure 2.
Protein architectures of full length BLUF and LOV photoreceptor-effector systems. Photoreceptor domains are colored according to secondary structure: helices, orange; β-sheets, blue. N- and C-terminal helical domain extensions are colored in light and dark red, respectively, and elements important for signal transduction are colored in purple. (A) AppA-PpsR system from Rhodobacter sphaeroides [27]. AppA (shown as cartoon) senses blue light using a BLUF domain and, via its C-terminal SCHIC domain, binds to a dimer of the transcriptional repressor PpsR (a truncated construct is shown in surface representation). Illumination alters the DNA-binding ability of PpsR through allosteric modulation of its helix-turn-helix motif (HTH; not present in the AppA-PpsR2 core complex structure—cf. Figure 6). (B) Blue-light regulated phosphodiesterase 1 (BlrP1) from Klebsiella pneumonia [86]. BlrP1 contains an N-terminal BLUF domain and a C-terminal EAL phosphodiesterase domain. The protein homo-dimerizes through a conserved interface made up of two dimerization helices and one compound helix formed by two short helices provided by each protomer (both shown in purple). Communication between the BLUF domains and the EAL active sites is bidirectional and mediated through the dimer interface [87]. (C) Light-activated transcription factor EL222 from Erythrobacter litoralis [14]. In contrast to the well-studied LOV2 from Avena sativa phototropin (shown in (E) as part of PA-Rac1), the C-terminal Jα helix (shown in red) does not pack against the LOV β-sheet. Instead it is replaced by a helix of the HTH DNA-binding motif. Illumination releases the HTH motif from the LOV β-sheet and thus enables DNA binding. (D) Light-activated monomeric histidine kinase EL346 from Erythrobacter litoralis [22]. Two α-helices of a dimerization/histidine phosphotransfer-like (DHpL) domain (shown in purple) pack against the LOV β-sheet and couple the photoreceptor to the catalytic/ATP-binding (CA) domain with bound adenosine 5′-(β, γ-imido)triphosphate (AMP-PNP, shown as stick model). Illumination is assumed to loosen the LOV-DHpL interactions, thus enabling CA domain activation. (E) Synthetic photoactivatable GTPase Rac1 (PA-Rac1) [19]. PA-Rac1 was generated by fusing the LOV2 domain from Avena sativa phototropin 1 to the small human Rac1 GTPase. A Ca2+ ion (purple sphere) bound in the interdomain interface restricts the dynamics of the Jα helix (dark red) packed against the LOV domain core. Illumination perturbs the Jα helical structure and renders the interface more dynamic, allowing the GTPase to engage in β-strand pairing with downstream effectors through its peripheral β-strand (purple) [83]. (F) Synthetic light-regulated histidine kinase YF1 [93]. The dimeric protein was generated by replacing the oxygen-sensing PAS domain of Bradyrhizobium japonicum FixL with structurally homologous LOV photoreceptor domain from Bacillus subtilis YtvA. The main part of the LOV dimer interface is formed by the N-terminal flanking helices (light red) that pack against the LOV β-sheet of the opposite subunit of the dimer. The C-terminal Jα helices (dark red) form a coiled coil structure extending away from the photoreceptor domains, connecting them to a dimerization/histidine phosphotransfer (DHp) domain (colored in purple) which forms an interaction platform for the CA domains. The activation mechanism is proposed to involve twisting of Jα upon illumination.