Fig. 2.
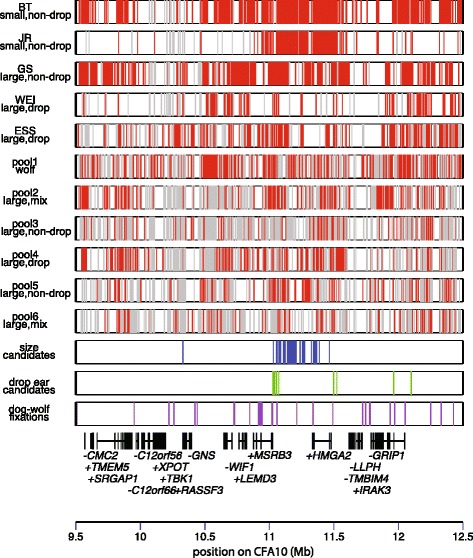
Patterns of SNP variation in a 3 Mb region on CFA10. The first 5 bars show variation in the sequence capture (SC) pools of single breeds and the next 6 bars show variation in the whole genome sequencing pools (WGS; see Table 2 for details). Red lines represent SNP positions that are fixed for a non-reference allele in a particular pool, grey lines represent SNP positions that cannot be confidently assessed due to low coverage. Sites that are polymorphic within a breed, or that match the reference allele are not marked. The bottom 3 bars represent SNPs that display patterns of fixation that matches phenotypic variation. Candidate SNPs for controlling variation in body mass (blue) ear type (green) and those that are fixed for alternate alleles in all dogs compared to wolves (purple) are shown. The location of protein coding genes in the region are also shown, which were identified by mapping human RefSeq genes onto the canFam2.0 dog assembly. Genes are labelled +/− according to direction of transcription. Ear and body mass candidates are concentrated in a region between the MSRB3 and HMGA2 genes, whereas a cluster of dog-wolf fixations is found within the MSRB3 gene
