Figure 5.
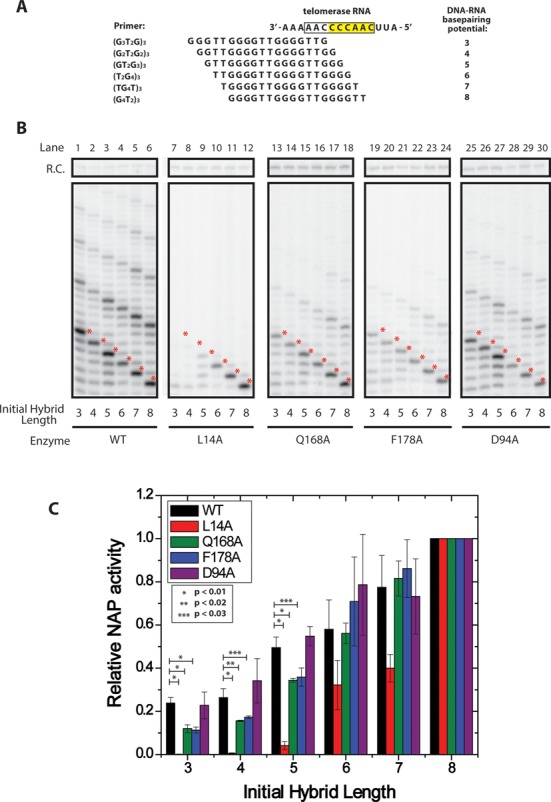
Telomerase activity assays demonstrate TEN domain mutations affect nucleotide addition processivity of primers with short RNA–DNA duplexes. (A) Primer permutants used in in vitro extension assays. Primers were length-matched at 3 telomeric repeats (18 nts), but staggered such that they formed different initial potential RNA–DNA duplex lengths with template RNA. (B) Telomerase was reconstituted in rabbit reticulocyte lysate and telomerase activity was assayed on the six DNA primers corresponding to six potential RNA–DNA hybrid lengths. WT enzyme was compared against enzyme harboring L14A, Q168A, F178A and D94A mutations. Mutants were assayed for primer-specific NAP defects by comparing the accumulation of the first repeat addition band (red asterisks) between primers tested with the same enzyme. (C) Quantification of relative NAP activity as a function of initial primer duplex length. Telomerase activity assay gels (Figure 5B) were performed in triplicate and quantified using the program SAFA (26). The quantification of each band was then corrected for specific activity. NAP was quantified as the amount of product that was extended to the 5′ end of the RNA template (Figure 5B, red asterisks, see Materials and Methods for details). For each wild-type or mutant enzyme, the observed NAP activity for each primer variant was normalized to the (GGGGTT)3 primer, which has the potential to form eight basepairs of RNA–DNA hybrid, and displayed the maximal NAP activity. P-values indicating statistical significance are as marked on the graph, error bars indicate one standard deviation based on triplicate measurements.
