Figure 1.
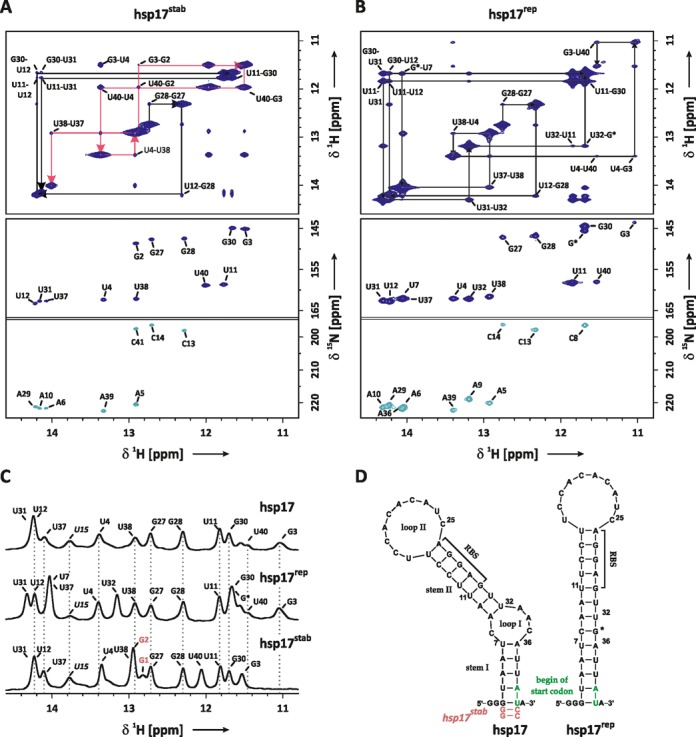
(A) Imino proton resonance assignment of the stabilized hsp17stab RNAT: imino region of a 1H-1H-NOESY spectrum (top) and HNN-COSY spectrum (below). Resonances of G2 and C41 originate from the mutation A40CC, which was introduced to stabilize stem I. Sequential walks for stems I and II are highlighted in red (G2-U38) and black arrows (G27-U31). Both experiments were recorded at T = 284 K. (B) Imino resonance assignment of the hsp17rep RNAT: imino region of a 1H-1H-NOESY spectrum (top) and HNN-COSY spectrum (below). Sequential walk is indicated by black arrows (G27-U40). Both experiments were recorded at T = 273 K. Base-pair numbering refers to the wild-type sequence shown in (D), G* denotes the mutation AAC(33–35)G. (C) Imino proton resonance assignment of hsp17 based on comparison of 1H-spectra of hsp17rep and hsp17stab recorded at 274 K. (D) Secondary structure of hsp17, hsp17rep and hsp17stab based on NMR resonance assignment. RBS denotes the ribosome binding-site.
