Figure 2.
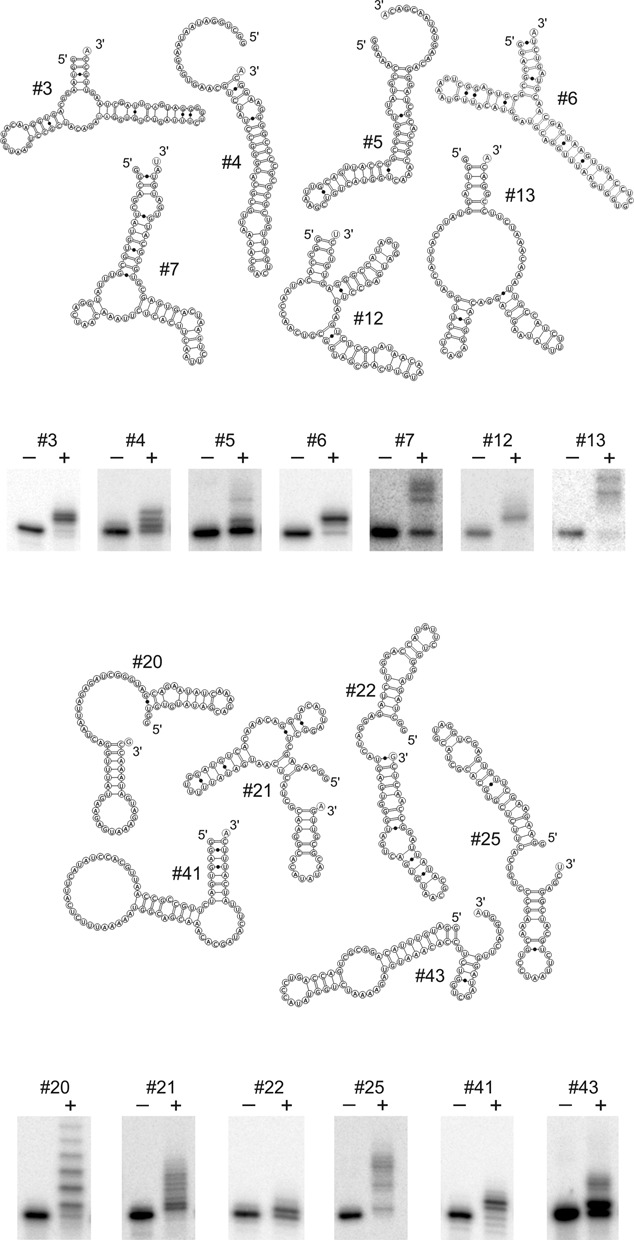
Individual candidate transcripts are true substrates for CCA addition. From the list of candidates presented in Figure 1, 13 RNA molecules were tested individually with the CCA-adding enzyme (+). The negative control represents the transcripts incubated without enzyme. All candidates show nucleotide additions with varying efficiency, ranging from the incorporation of a single nucleotide (partial CCA end, #22) to the addition of three or more residues. The secondary structure models show that most of the candidates carry base-paired 5′- and 3′-ends with 3′-terminal nucleotide overhangs, corresponding to an acceptor stem-like structure. Yet, also single-stranded 3′-ends are tolerated for nucleotide addition, although at a rather low efficiency (#5, #25, #43).
