Figure 2.
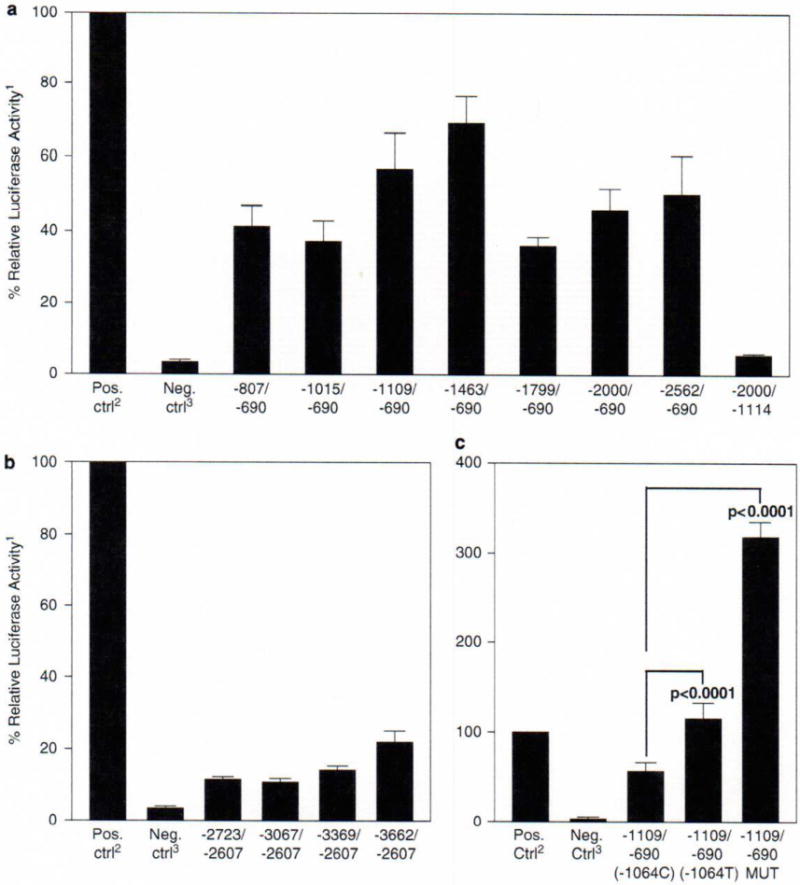
Luciferase assays. (a) Luciferase expression for transcriptional start site one constructs. A total of 5 × 104 HeLa cells were grown in 24-well plates in 0.5 ml media (Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum, glutamine, penicillin and streptomycin) overnight. The HeLa cells were then transfected with 400 ng of luciferase reporter constructs and 100 ng of pRL-TK Vector (Promega Corporation) using FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN, USA). After 48 h incubation, passive lysis of the cells was performed and the lysate was used to measure the luciferase expression using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer’s protocol. A GENios Pro (Tecan Systems Inc., San Jose, CA, USA) was used to measure the relative light units. (b) Luciferase expression for transcriptional start site two constructs. Cell culture, transfection and luciferase assays were performed as described in (a). (c) Luciferase expression of −1064C/T and Sp1 mutant. Cell culture, transfection and luciferase assays were performed as described in (a). To mutate the Sp1 site located at −1073 upstream of the translation start site, the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) was used according to the manufacturer’s protocol. This Sp1 site was mutated from 5′-CCGCCC-3′ to 5′-CCGAAC-3′ using the following primers: QCPro_mSP1F (5′-TCCTGACCTCAGGTGATCCGAACACCTCGGCCTCCCAAAGTG-3′) and QCPro_mSPlR (5′-CACTTTGGGAGGCCGAGGTGTTCGGATCACCTGAGGTCAGGA-3′). The -1109/-690 wild-type luciferase reporter construct was used for the template DNA. The Sp1 mutant construct was verified by direct sequencing. 1All luciferase activities are expressed as a percentage of the positive control, with the positive control as 100%. 2pGL3-Control Vector (Promega Corporation) was used as the positive control. 3pGL3-Enhancer Vector (Promega Corporation) without an insert was used as the negative control. P-values were determined by Student’s t-test; n = 12 for all assays except controls (n = 15) and −1109/−690 MUT (n = 6). Error bars represent the standard error of the mean.
