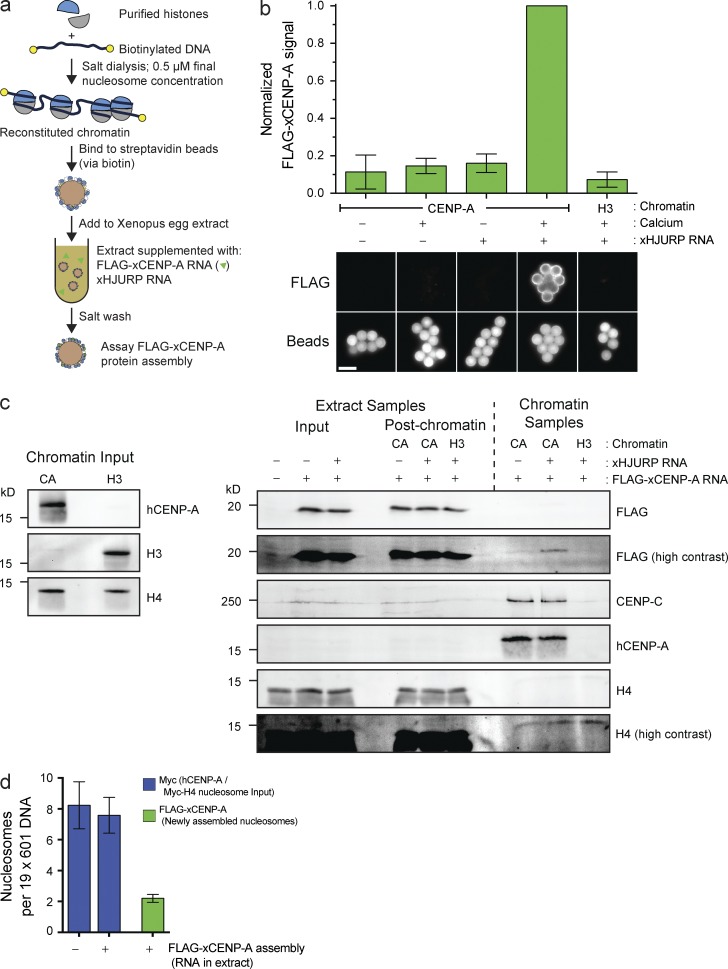
Figure 1.
Reconstitution of CENP-A assembly in vitro. (a) Schematic of in vitro CENP-A assembly assay. (b) In vitro FLAG–xCENP-A assembly requires HJURP and mitotic exit. Extracts were supplemented with calcium, xHJURP RNA, or both. The top graph shows the means ± SEM; n = 5. The bottom images show FLAG–xCENP-A staining (FLAG) and bead autofluorescence (Beads). Bar, 5 µm. (c) Characterization of FLAG–xCENP-A assembly. (left) The levels of CENP-A (hCENP-A), histone H3, and histone H4 on the beads before extract addition. (right) The levels of FLAG–xCENP-A, CENP-C, and histone H4 in the extract before chromatin bead addition and after bead recovery (Extract Samples), and the chromatin bead–bound proteins after their recovery from the extract (Chromatin Samples). (d) Summary of quantitative Western blots (see Fig. S1 c) estimating the number of input nucleosomes (Myc-H4 signal) and newly assembled FLAG–xCENP-A nucleosomes per chromatin array; bars show the means ± SEM; n = 5.