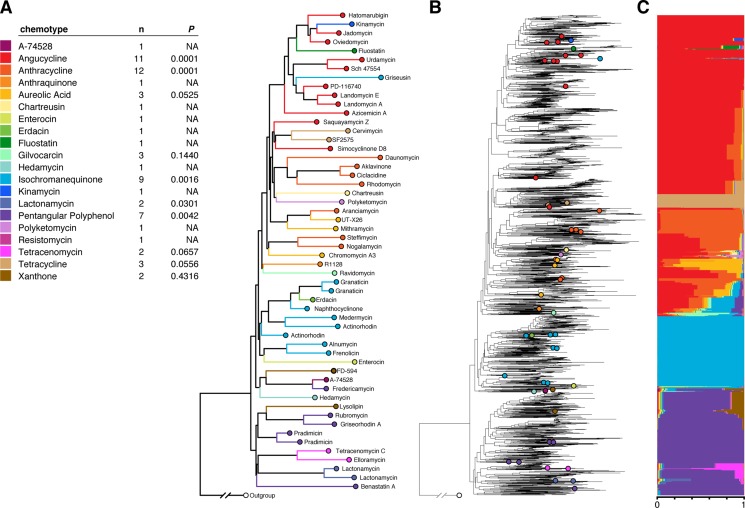
Fig 2. Estimating putative antibiotic production.
A) Phylogeny of reference sequences, obtained by pruning environmental sequences from B. Tips are labeled by the polyketide produced and colored by polyketide chemotype. n denotes the number of reference sequences of the given chemotype, and P reflects the clustering of the chemotype on the phylogeny (computed as a z-score, see Material and Methods). The clustering is significant or marginally significant for almost all chemotypes for which it could be computed. B) The phylogeny of environmental KSα genes (black), along with reference sequences (colored), allows estimating for each environmental sequence and each chemotype the probability that the sequence codes for the chemotype. C) Each colored stripe indicates the inferred probability that the corresponding sequence codes for the chemotype represented by the color.