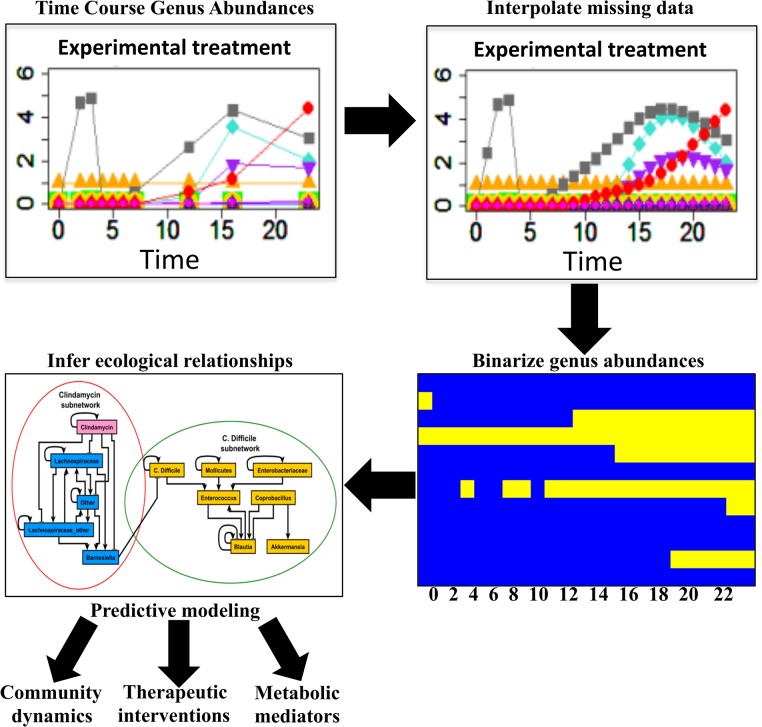
Fig 1. Dynamic analysis workflow.
Time course genus abundance information was acquired from metagenomic sequencing of mouse gastrointestinal tracts under varying experimental conditions. Missing time points from experimental data were estimated such that genus abundances existed at the same time points across all treatment groups. Next, genus abundances were binarized such that Boolean regulatory relationships could be inferred. A dynamic Boolean model was constructed to explore gut microbial dynamics, therapeutic interventions, and metabolic mediators of bacterial regulatory relationships.