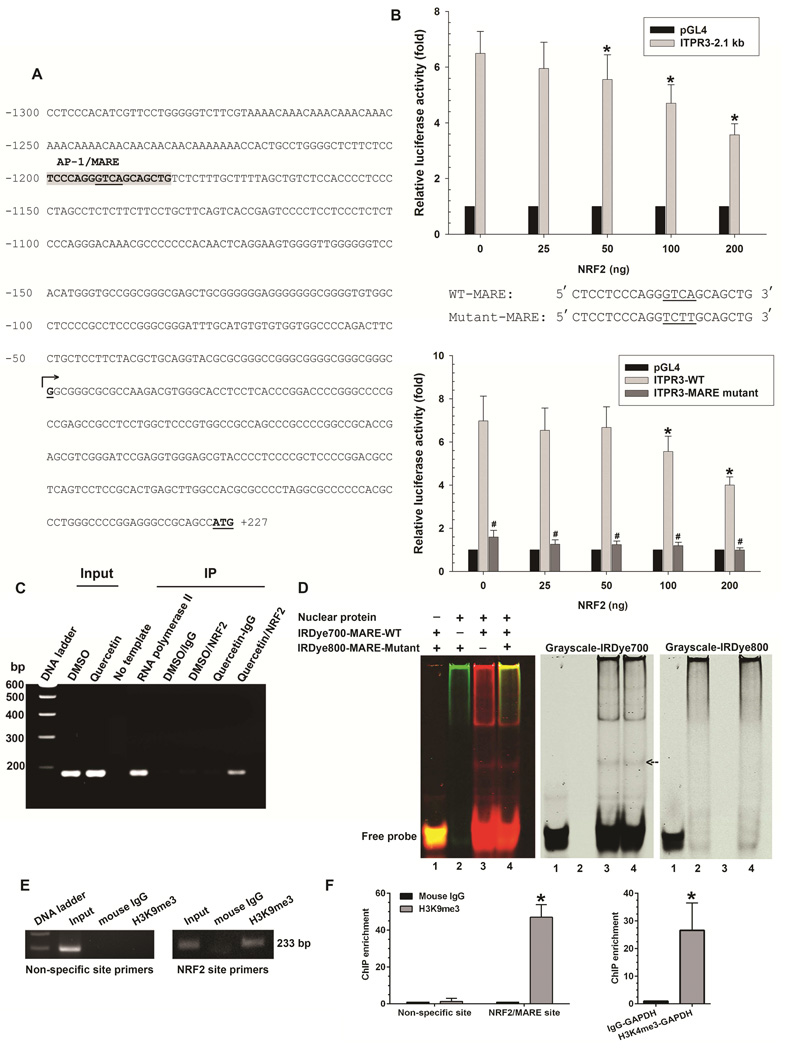
Figure 1. NRF2 binds to MARE on the human ITPR3 promoter and suppresses promoter activity.
(A) DNA sequence of the proximal (2.1 kb) region of the human ITPR3 promoter. Potential transcriptional binding sites for NRF2/MARE are shaded in gray, and underlined nucleotides are the core sequence of the NRF2/MARE. Numbered positions refer to the transcription starting sites (nucleotide, +1, arrow). (B) NRF2 inhibits ITPR3 promoter activity in a dose- and MARE-dependent fashion. Upper panel shows gene reporter assays. The ITPR3 promoter construct (p-1812/+326-Luc) was co-transfected with the NRF2 expression plasmid in NHC cells. Lower panel shows luciferase activity of the MARE mutant. NHC cells were co-transfected with mutated core region of MARE in p-1812/+326-Luc and its corresponding wild-type constructs with different amounts of NRF2. Results are presented as fold changes relative to empty luciferase vector (pGL4) from the same dual luciferase assay experiment. *p<0.05 relative to empty vector (n=4 separate transfection experiments). (C) ChIP assay. Chromatin prepared from NHC cells treated with DMSO or quercetin (Input) was immunoprecipitated with NRF2 antibody. Precipitated chromatin DNA was detected by a primer set covering the MARE region using PCR. RNA polymerase II was used as a positive control. Normal rabbit IgG was used as a negative control. (D) Electrophoretic mobility shift assay. Nuclear extract isolated from NHC cells was incubated with either IRDye700-labeled oligonucleotides corresponding to the wild-type MARE motif (IRDye700-MARE-WT) or IRDye800-labeled oligonucleotides corresponding to the mutated MARE motif (IRDye800-MARE-Mutant). Analysis of direct binding was conducted as indicated. Black arrow indicates positions of the DNA–protein complexes. (E) Representative image showed H3K9 methylation of histone H3 surrounding the NRF2/MARE site. Chromatin was prepared from NHC cells transfected with NRF2 plasmid. Chromatin was immunoprecipitated with anti-trimethyl H3K9 antibody or normal mouse IgG. Precipitated chromatin DNA with H3K9me3 or normal mouse IgG was detected by a primer set covering the specific NRF2/MARE and non-specific regions using PCR. (F) Quantification of ChIP enrichment of H3K9 methylation. Normal mouse IgG served as a control. Negative control consisted of amplification of a non-specific site (3'-UTR region of ITPR3 gene) distant from the promoter region. NHC cells transfected with NRF2 plasmid were assayed for H3K9 methylation by ChIP with primers specific for the MARE region on the ITPR3 promoter. Enrichment was quantified by qPCR. Positive controls were immunoprecipitated with anti-trimethyl H3K4 antibody. Precipitated chromatin DNA with H3K4me3 was detected using primers for GAPDH promoter. Increased H3K4me3 signal was consistent with constitutive activation of the GAPDH gene. Results are expressed as the fold-increase of the ratio to the normal mouse IgG control. Values are mean ± SD, n=6 (*p<0.005).