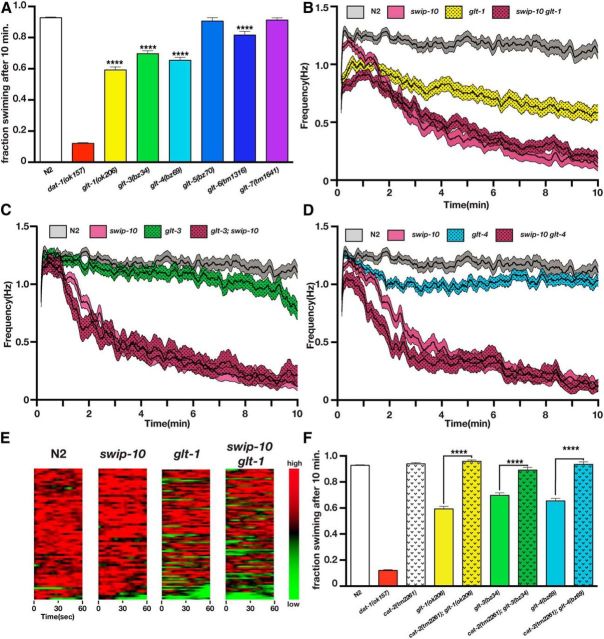
Figure 7.
Loss of GLT genes produces DA-dependent Swip. A, Manual Swip assays of glutamate transporter alleles. Loss of glt-1, glt-3, glt-4, or glt-6 produces a significant Swip phenotype relative to N2 when assessed at a single 10 min time point. B, Automated thrashing analysis reveals that loss of glt-1 produces a significant Swip phenotype relative to N2 (significant reduction at all time points). Multiple comparisons of swip-10 versus swip-10 glt-1 mutants show that swip-10 glt-1 mutants are significantly reduced from swip-10 mutants from time points 0–48 s. C, Automated thrashing analysis reveals that loss of glt-3 produces a significant Swip phenotype relative to N2 (significant reduction at scattered time points during minutes 6–9 from 9:13–10:00). Multiple comparisons of swip-10 versus glt-3;swip-10 mutants show that glt-3;swip-10 mutants are significantly reduced from swip-10 mutants from time points 1:12–1:18, 1:29–1:32, and 1:34–1:39. D, Automated thrashing analysis reveals that loss of glt-4 produces a significant Swip phenotype relative to N2 (significant reduction at scattered time points during minutes 2–6). Multiple comparisons of swip-10 versus swip-10 glt-4 mutants show that swip-10 glt-4 mutants are significantly reduced from swip-10 mutants from time points 0:55–1:31, 1:37–2:01, and 2:07–2:19. E, Heat maps of the first minute of automated recordings for N2, swip-10, glt-1, and swip-10 glt-1 strains. Colors represent normalized thrashing values within that strain where red is the highest thrashing value and green is no movement. F, Swip in glt-1, glt-3, and glt-4 is DA dependent because loss of cat-2 restores swimming behavior in glt-1, glt-3, and glt-4 to wild-type swimming levels. For A and F, data were analyzed using one-way ANOVA with multiple Bonferroni posttests where ****p < 0.0001 and error bars represent SEM. For B–D, single worm recordings were performed as described in the Materials and Methods and analyzed with SwimR. Data were analyzed by two-way ANOVA with multiple Bonferroni posttests at each time point. n ≥ 33 for each strain.