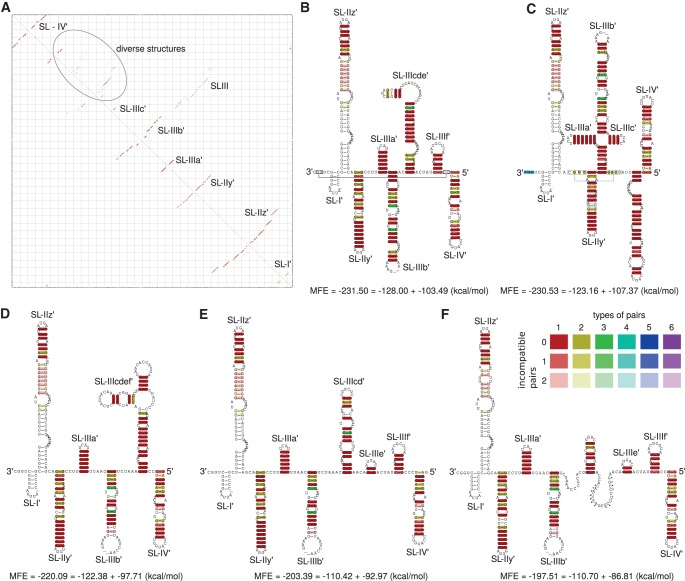
FIGURE 6.
Alignment based secondary structure of the 3′ end of the minus-strand with 86 isolates. (A) RNAalifold base-pairing probability matrix. The upper half of the matrix shows all base pair probabilities <10−6. The lower half shows the base pair probabilities of the MFE structure. (B) Consensus secondary structure without constraints. (C) Consensus secondary structure with constraints: The last 3 nt at the very 3′ end were forced to be unpaired (blue). The resulting structure mirrors the secondary structure of the domain SLIII from RNA plus strand. (D) Consensus secondary structure with constraints forcing the postulated structure of Smith et al. (2002). (E) Consensus secondary structure with constraints forcing the postulated structure of Dutkiewicz et al. (2008). (F) Consensus secondary structure with constraints forcing the postulated structure of Schuster et al. (2002). (Dots) Nucleotides base-pairing with an extended version of the 3′ end; for details see Schuster et al. (2002). (Connected rectangles) Further interactions calculated by LocARNA. White base-pairings are derived from isolates being sequenced only partially.