FIGURE 3.
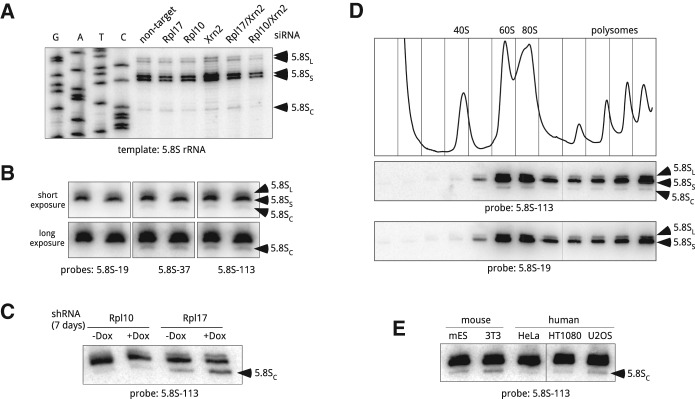
Heterogeneity of 5.8S rRNA in ribosomes from mouse and human cells. (A) Primer extension analysis of the 5′ ends of gel-purified 5.8S rRNA using primer 5.8S-113. Cells were analyzed 48 h after transfections with the indicated siRNAs. (B) A shortened form of 5.8S rRNA is detectable by northern hybridizations. Duplicate samples of RNA from growing 3T3 cells were resolved on a polyacrylamide/urea gel, transferred to a nylon membrane and hybridized with the indicated probes. Probes 5.8S-37 and 5.8S-113 detect all three forms of 5.8S, whereas 5.8S-19 does not hybridize with 5.8SC; see Figure 2A for probe positions. (C) Prolonged depletion of Rpl17, but not Rpl10, increases the amount of 5.8SC rRNA in mouse cells. Cells harboring doxycycline-inducible shRNAs were incubated with doxycycline for 7 d or remained untreated (+Dox, −Dox). RNA from these cells was separated by polyacrylamide gel electrophoresis and analyzed by Northern blotting. (D) 5.8SC rRNA is incorporated into ribosomes that are translationally competent. Growing 3T3 cells were lysed and cytoplasmic ribosomes were separated by centrifugation through 10%–45% (w/w) sucrose gradients. The absorbance profile of the gradient at 254 nm is shown at the top. RNA isolated from each gradient fraction was analyzed by polyacrylamide gel electrophoresis and northern hybridizations with probes that either detect (5.8S-113) or do not detect (5.8S-19) the 5.8SC isoform. (E) 5.8SC rRNA is formed in different mouse and human cell lines. RNA was extracted from the indicated mouse and human cell lines, and analyzed as in C. (mES) Mouse embryonic stem cells.
