FIGURE 4.
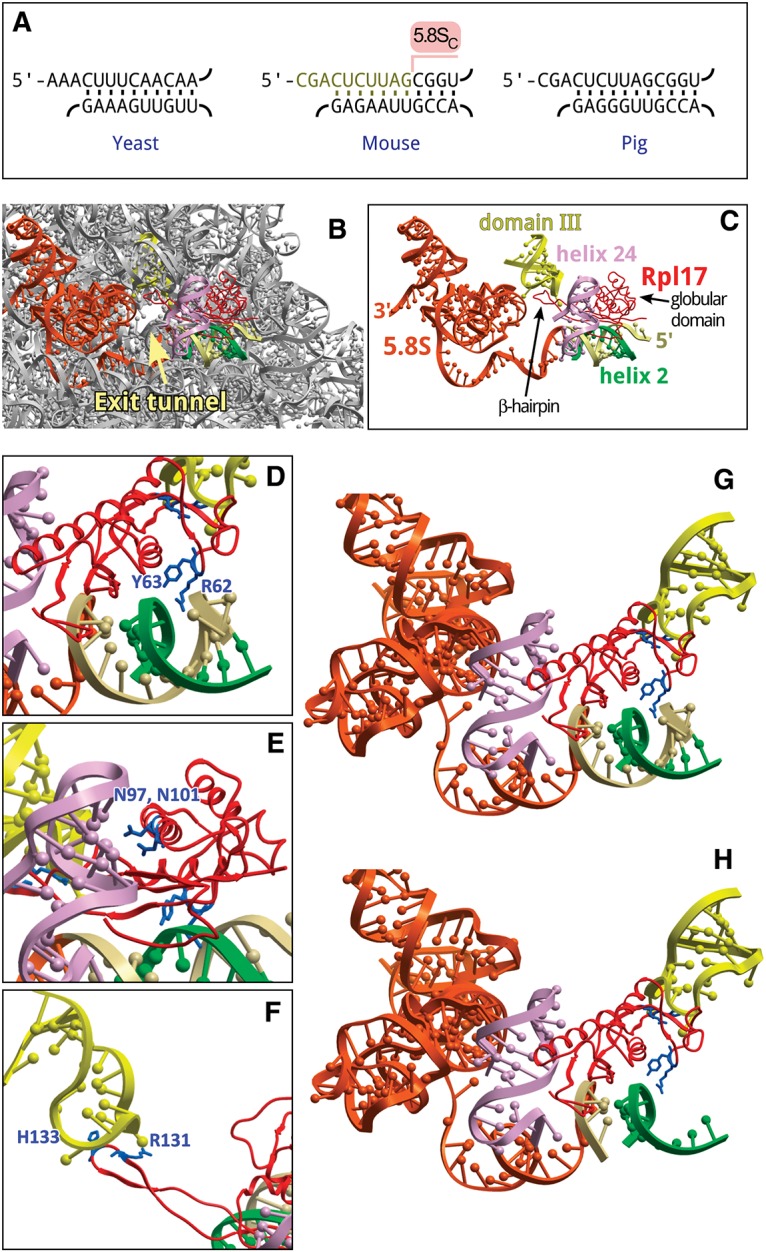
Structural implications of the 5′ truncation in 5.8S rRNA. (A) Conservation of helix 2 in the large subunit rRNA of Saccharomyces cerevisiae, Mus musculus, and Sus scrofa. Top strand, 5.8SS rRNA; bottom strand, 25/28S rRNA. Ten nucleotides missing in 5.8SC rRNA are indicated in the mouse sequence. (B) Position of 5.8S rRNA, helix 2 and Rpl17/uL22 relative to the exit tunnel in the 60S subunit of S. scrofa (PDB: 3J71). (C) The same view showing only structural elements that surround the exit tunnel and interact with Rpl17 (red). Orange, 5.8S rRNA, the first 14 nucleotides at the 5′ terminus shown in beige. Green, helix 2, domain I (28S rRNA nucleotides 419–430, 5′-accguuaagag-3′); light purple, helix 24, domain I (28S rRNA nucleotides 388–410, 5′-aacuuugaagagagaguucaaga-3′); yellow, domain III hairpin loop (28S rRNA nucleotides 1587–1606, 5′-guccugacgugcaaaucggu-3′). (D–F) Rpl17 residues that contact helix 2, helix 24, and the loop in domain III, respectively. (G,H) Residues of the Rpl17 globular domain facing intact or cropped helix 2, respectively. Images were generated using ICM-Browser (Molsoft).
